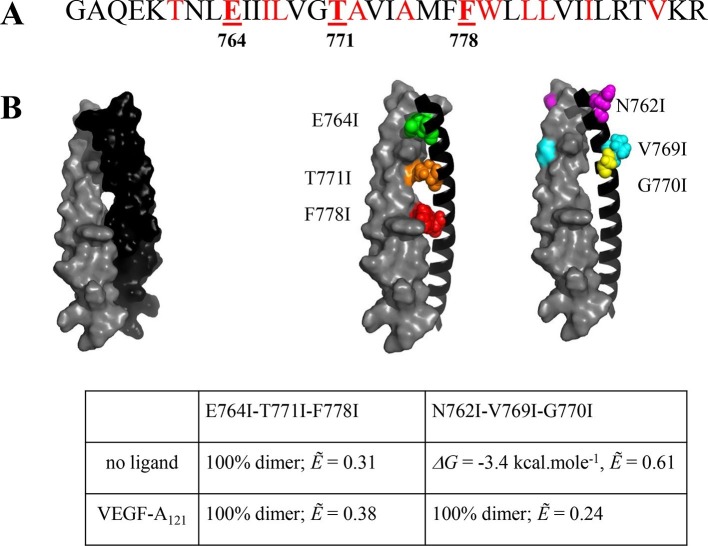
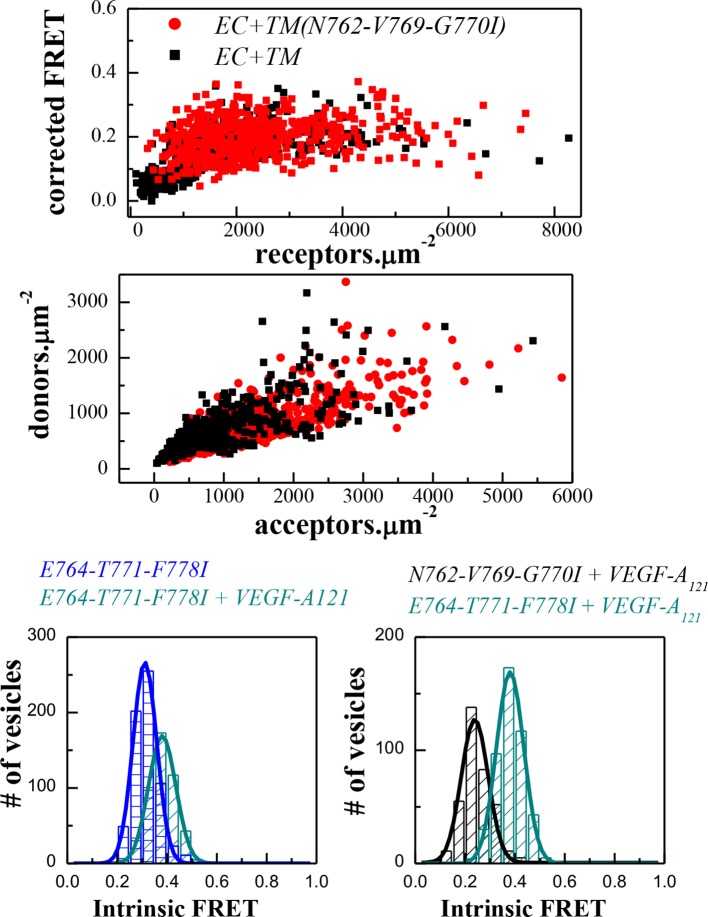
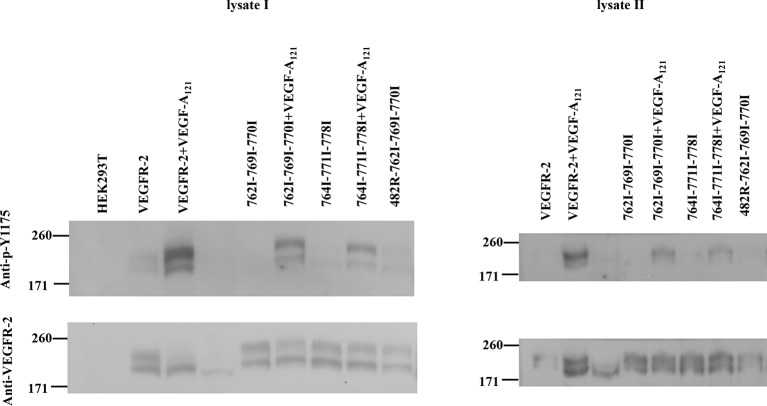
Figure 4. The published NMR structure of the isolated VEGFR-2 TM domain (Manni et al., 2014b) is consistent with the unliganded TM dimer structure observed in the FRET experiments.
(A) Amino acid sequence of wild-type VEGFR-2 TM domain. The amino acids that mediate helix-helix contacts in the NMR dimer structure (Manni et al., 2014b) are shown in red. (B) The NMR structure, with two sets of amino acids highlighted. Left: a space-fill model of the wild-type VEGFR-2 TM dimer, based on NMR experiments. Center: E764, T771, and F778 help mediate helix-helix contacts in the NMR structure (Manni et al., 2014b) (also shown bold and underlined in (A)). Right: N762, V769, and G770 face away from the dimer interface, into the membrane. Two sets of mutations: a E764I-T771I-F778I set and a N762I-V769I-G770I set, were engineered. Table: Results of FRET experiments for the two sets of mutants (see also Table 1). The dimerization of the unliganded EC+TM construct was affected by the E764I-T771I-F778I but not by the N762I-V769I-G770I set of mutations, suggesting that the unliganded VEGFR-2 TM dimer is stabilized by contacts involving E764, T771, and/or F778, as in the NMR structure. At least one of the amino acids N762, V769, and G770 forms direct intermolecular contacts in the TM domain in the ligand-bound state.