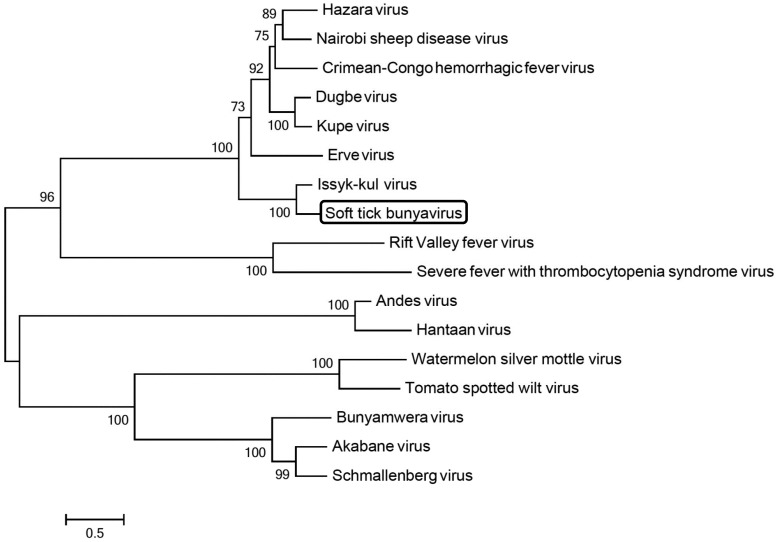
Fig. 2.
Maximum likelihood tree based on nucleotide sequence of viruses of the family Bunyaviridae. The percentage of replicate trees in which the associated taxa are clustered together in the bootstrap test (1,000 replicates) was calculated. The length of the bar corresponds to the degree of sequence divergence. Phylogenetic analyses were conducted in MEGA 6. (Accession No.; CCHFV GU477492.1, Hazara virus DQ076419.1, NSDV EU697951.1, Kupe virus EU257628.1, Dugbe virus U15018.1, Erve virus JF911697.1, Issyk_Kul virus KF892055.1, Rift valley virus JF311376.1, SFTSV JQ684871.1, Hantaan virus JQ083393.1, Schmallenberg virus KC355457.1, Akabane virus AB190458.1 and Tomato spotted wilt virus KC261962.1)