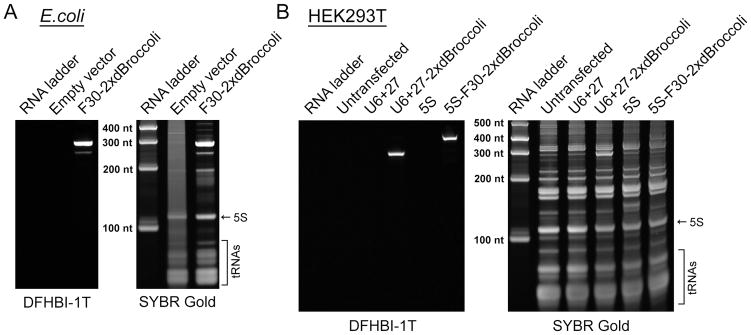
Figure 2.
Broccoli expression is readily detected using the in-gel staining protocol. (A) Bacterial cells were transformed with either pET28c plasmid (negative control) or with pET28c-F30-2xdBroccoli and the RNA was expressed for four hours. Then total RNA was isolated, separated using denaturing PAGE and stained with DFHBI-1T and then with SYBR Gold, as described the Basic Protocol 1. DFHBI-1T specifically reveals Broccoli-containing bands while SYBR Gold detects total cellular RNA. (B) Mammalian (HEK293T) cells were either untransfected or transfected with pAV5S or pAVU6+27 (negative controls) or with pAV5S-F30-2xdBroccoli or pAVU6+27-F30-2xdBroccoli. After 72 h expression total RNA was isolated, separated using denaturing PAGE and stained with DFHBI-1T and then with SYBR Gold, as described in the Alternate Protocol 1. Broccoli-containing bands are clearly detectable even in such a complex mixture as total mammalian cell RNA.