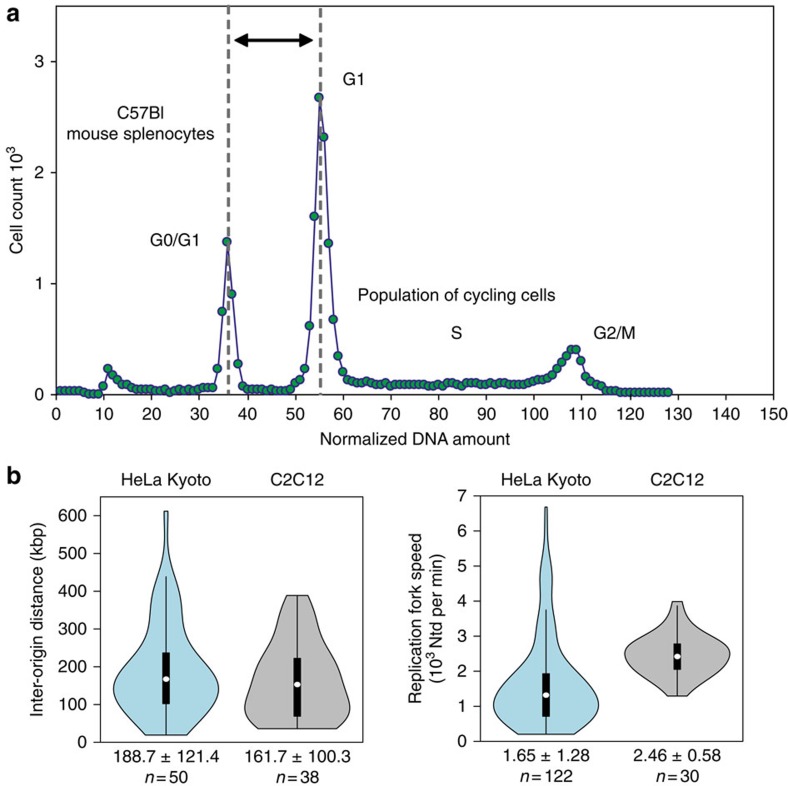
Figure 2. Genomic DNA content and DNA fibre analysis of replicons.
(a) DNA flow cytometry histogram of ethidium bromide/olivomycin stained HeLa Kyoto mCherry-PCNA expressing cells admixed with C57Bl mouse splenocytes is shown. The peak at channels 34–38 corresponds to the G1/G0 peak of non-cycling splenocytes. HeLa Kyoto cell cycle distribution is represented by a typical DNA flow cytometry histogram consisting of G1, S-phase and G2/M populations. To calculate the amount of genomic DNA in the cycling cell line, G1/G0 peak of mouse splenocytes and G1 peak of the cell line were approximated with Gaussian distributions and the relative position of the G1 peak was calculated (for details see methods and Table 2). (b) Cells were pulse labelled with IdU for 30 min, followed by a 30 min CldU pulse. Whole-genome DNA was extracted under gentle conditions and single DNA fibres were stretched with the constant factor of 2 kbp per μm. Incorporated nucleotides were immunostained and signals acquired in a wide-field microscope. Fluorescent tracks were measured by hand and used to calculate mean IOD and RFS. For details see methods; Supplementary Fig. 2 and Tables 3 and 4.