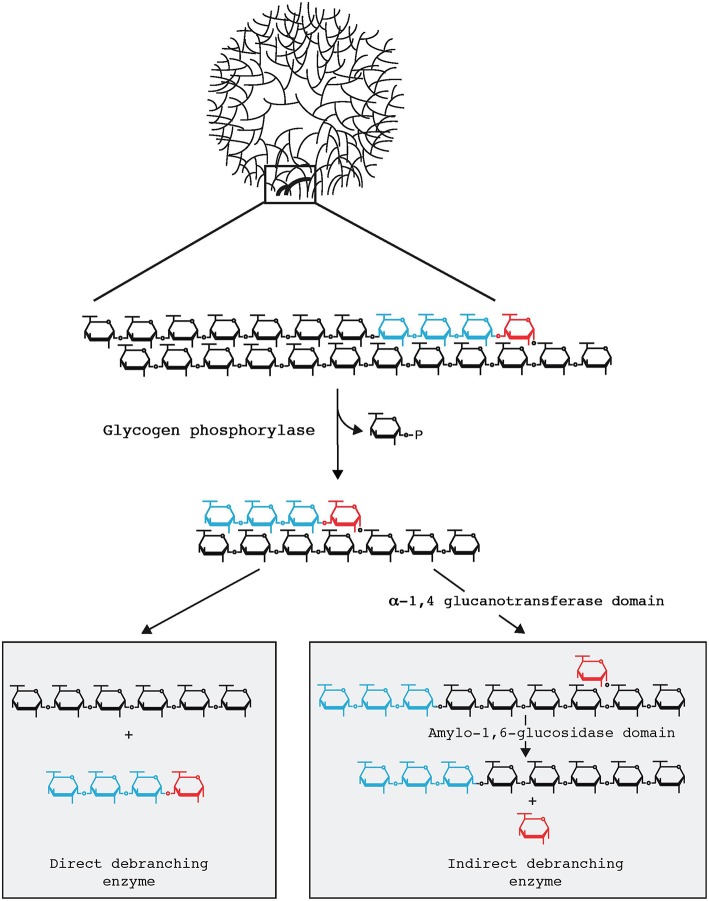
Figure 1.
Glycogen particle debranching in eukaryotes and bacteria. A 40 nm diameter hydrosoluble glycogen particle is displayed on top emphasizing the outer chains connected to the particle through a single α-1,6 linkage. One-third in weight of the total glucose resides are these outer chains which are directly available to the hydrosoluble enzymes involved in glycogen breakdown. In both bacteria and eukaryotes these outer chains enlarged in the top panel are first recessed through glycogen phosphorylase which in the presence of orthophosphate releases glucose-1-P. All reported glycogen phosphorylases stop at a four glucose residue distance from the first encountered α-1,6 branch (displayed in red). Bacteria and eukaryotes differ at the subsequent steps (Cenci et al., 2014). Bacteria directly hydrolyse the remaining outer α-1,6 linkage through the so-called direct debranching enzyme (DBE) generating a smaller glycogen particle and releasing a small chain of four glucose residues (maltotetraose) in the cytosol. Bacteria must then metabolize these small chains and do so with the help of MOS metabolism enzymes. DBE belongs to the GH13 glycosyl hydrolase family. Eukaryotes first hydrolyse the α-1,4 preceding the branch and transfer the maltotriose (displayed in blue in the right panel) outer chain to the neighboring chain allowing for further digestion with glycogen phosphorylase. The α-1,6 linked glucose at the remaining branch is then hydrolyzed by a second active site on the same enzyme named indirect debranching enzyme (iDBE) releasing glucose which is phosphorylated to glucose-6-P by hexokinase. No MOS is released in the cytosol of eukaryotes through glycogen metabolism. iDBE is derived from a gene fusion between a GH13 and GH133 domain. Consequently one finds iDBE candidate sequences in all eukaryotes unrelated to Archaeplastida that accumulate glycogen [all opisthokonts (fungi and animals), amoebozoa, alveolata, glycogen accumulating excavata (Trichomonas, Giardia)] whereas the fusion has never been seen in bacteria despite the extensive databases that are available (Ball et al., 2011, 2015). Bacterial GH13 DBE is likewise never seen in the aforementioned eukaryotic clades unrelated to Archaeplastida. In addition eukaryotes do not contain cytosolic enzymes of MOS metabolism with the exception of maltase (DPE2) which selectively hydrolyses maltose generated by the eukaryotic specific enzyme β-amylase.