Fig. 1.
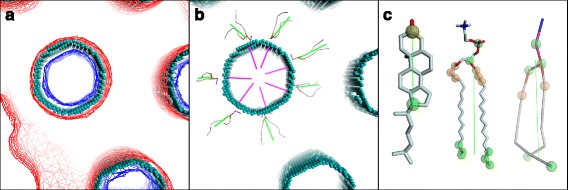
Lipid and water densities and interface. (a) Illustration of the determination of the lipid water interface for a DOPE (dioleoylphosphoethanolamine)/Cholesterol inverted-hexagonal phase [20]. The lipid density map is shown in red, the solvent density in blue and the interface as cyan spheres. (b) Same system as (a), showing the interface in cyan spheres with several normal vectors shown in magenta and DOPE lipids with their director vectors shown as in (c). (c) Examples of selections used as headgroup, tail, and to calculate distances between lipids. Atoms used to define the director vector (headgroup and tail) are shown as green translucent spheres, atoms used in distance calculation as orange, and atoms used for both are in khaki. Director vectors are shown as green lines. From left to right, we show an all-atom cholesterol and DPPC (dipalmitoylphosphocholine) lipid (CHARMM force-field [36]) and a coarse-grained DOPE lipid (Martini force-field [37]). For phospholipids we typically use the last three carbon atoms of each acyl chain as the tail position, whereas we use several atoms from the hydrophilic region to define the head group (for example the phosphate atom and the first carbon atom linking the two acyl chains). For cholesterol we use the carbon of the carboxyl group and the last carbon of the rigid aromatic region of the molecule as head and tail respectively. Atoms used for distance calculations should correspond to atoms lying on the pivotal plane and hence depend on the system studied, but typically the pivotal plane is situated just below the hydrophilic region of the membrane and hence, for phospholipids, the first few carbon atoms of the acyl chains can be used. The corresponding dictionaries for the examples given here are: tail_dict={'CHL1': 'aname=C17', 'DPPC': 'aname=C214, C215, C216, C314, C315, C316', 'DOPE': 'aname=C5A, C5B'}, head_group_dict={'CHL1': 'aname=C3', 'DPPC': 'aname=P, C2', 'DOPE': 'aname=PO4, GL1, GL2'} and distance_dict={'CHL1': 'aname=C3', 'DPPC': 'aname=C22, C21, C23, C31, C32, C33', 'DOPE': 'aname=C1A, C1B'}
