Figure 1.
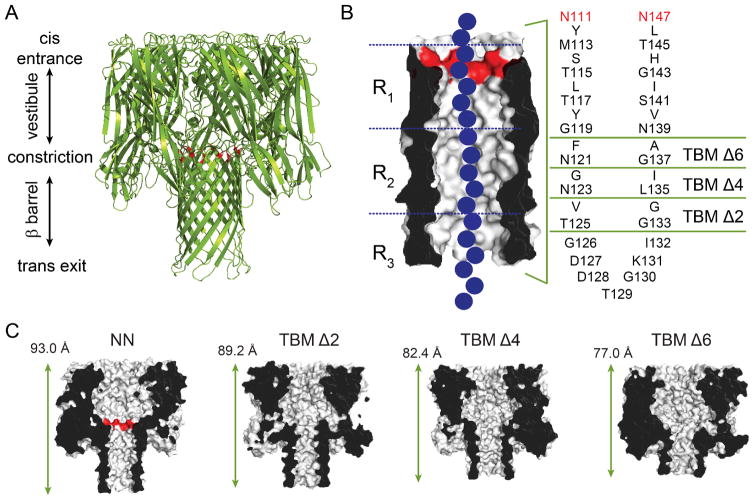
The α-hemolysin (αHL) protein nanopore. (A) Cartoon representation of the αHL pore (pdb: 7AHL). The αHL protein forms heptameric nanopores in lipid bilayers. The pore consists of an upper cap domain which contains a roughly spherical, water-filled vestibule, and a transmembrane domain. The E111N and K147N mutations15 at the top of the β barrel are highlighted in red, which replace the charged residues (Glu and Lys) at the central constriction. (B) The transmembrane domain is a 14-stranded, antiparallel β barrel. Each of the seven protomers contributes a pair of adjacent β strands (separated by a turn sequence; amino acids 126–132) to the barrel. The amino acid sequence of the transmembrane portion of the β strands for the most part alternates between hydrophilic residues (which face inwards towards the water-filled lumen of the pore) and hydrophobic residues (which face outwards towards the hydrocarbon core of the bilayer). To truncate the β barrel, rings of inward and outward facing residues from each of the two strands were deleted by PCR mutagenesis (leaving the turn sequence intact) to form truncated barrel mutant (TBM) proteins.11 The three nucleobase recognition sites within the β barrel, R1, R2 and R3, are also indicated. The three nucleobase recognition sites within the β barrel, R1, R2 and R3, are also indicated15, 16, 19 (C) Cut-through representations of the truncated mutants, TBMΔ2, Δ4 and Δ6, used in the present study. The length indicated is the distance between the Cα atoms of amino acids N17 (located at the top of the cap domain), of the 3rd subunit, and T129 (located at the bottom of the transmembrane domain), of the 7th subunit.11