Abstract
Pregestational diabetes significantly increases the risk of neural tube defects (NTDs). Maternal diabetes activates an Apoptosis Signal-regulating Kinase 1 (ASK1)-initiated pathway, which triggers neural stem cell apoptosis of the developing neuroepithelium leading to NTD formation. How high glucose of diabetes activates ASK1 is still unclear. In this study, we investigated the mechanism underlying high glucose-induced ASK1 activation. High glucose suppressed miR-17 expression, which led to an increase in its target gene Txnip (Thioredoxin-interacting protein). High glucose-increased Txnip enhanced its binding to the ASK1 inhibitor, thioredoxin (Trx), and thereby sequestered Trx from the Trx-ASK1 complex. High glucose-induced ASK1 activation and consequent apoptosis were abrogated by either the miR-17 mimic or Txnip siRNA knockdown. In contrast, the miR-17 inhibitor or Txnip ectopic overexpression mimicked the stimulative effect of high glucose on ASK1 and apoptosis. Thus, our study demonstrated that miR-17 repression mediates the pro-apoptotic effect of high glucose, and revealed a new mechanism underlying ASK1 activation, in which decreased miR-17 removes Trx inhibition on ASK1 through Txnip.
Keywords: high glucose, maternal diabetes, teratogenicity, ASK1 activation, miR-17, apoptosis
Diabetic pregnancy is associated with a high rate of birth defects including neural tube defects (NTDs) and congenital heart defects (CHDs), which have become a significant public health problem (Correa et al., 2008). Studies from our group (Gabbay-Benziv et al., 2015; Gu et al., 2015b; Li et al., 2012, 2013; Wang et al., 2015a,b,c,e,f; Wu et al., 2015; Yang et al., 2013, 2015) and others (Gareskog et al., 2007; Li et al., 2005; Pavlinkova et al., 2009; Salbaum and Kappen, 2010) have demonstrated that oxidative stress-induced pro-apoptotic kinase signaling plays a vital role in the induction of NTDs and CHDs in diabetic pregnancies. Specifically, the apoptosis signal-regulating kinase 1 (ASK1)-initiated pathway triggers neural stem cell apoptosis in the developing neuroepithelium of embryos exposed to maternal diabetes (Yang et al., 2013). In diabetic pregnancies, phosphorylation of ASK1 at Thr845 is increased in the developing neuroepithelium (Yang et al., 2013). Phosphorylation of ASK1 at Thr845 is essential for the catalytic activity of ASK1 and the induction of apoptosis (Tobiume et al., 2002). ASK1 activation stimulates the activity of the transcription factor FoxO3a, which increases the abundance of an apoptosis-promoting factor TRADD, therefore leading to caspase 8 dependent neuroepithelial cell apoptosis and finally NTD formation (Yang et al., 2013). Ask1 gene deletion inhibits maternal diabetes-induced neuroepithelial cell apoptosis and the development of NTDs (Yang et al., 2013). Thus, ASK1 activation is a causal event for maternal diabetes-induced NTD formation. However, the mechanism underlying maternal diabetes-induced ASK1 activation is still unclear.
MicroRNAs (miRNAs) are small endogenous non-coding RNAs that repress gene expression by binding to the 3′-untranslated region (3′-UTR) of mRNAs with imperfect complementation, resulting in direct translational repression, mRNA destabilization, or a combination of the 2 (Bartel, 2009; Lee et al., 1993; Xiao et al., 2011; Zhuang et al., 2013). The functions of miRNAs were originally described during normal development (Lee et al., 1993). Multiple lines of indirect evidence implicate that miRNAs mediate the adverse effect of high glucose on neural stem cells in the developing neuroepithelium and consequent NTD formation. First, miRNA profiling exhibits dynamic changes during mouse embryonic neurulation (Mukhopadhyay et al., 2011). Second, altered circulating miRNA profiling is observed in human pregnancies associated with NTDs (Gu et al., 2012, 2015b). Third, oxidative stress, a central causal event in diabetic embryopathy, modulates miRNA expression (Gu et al., 2015b; Magenta et al., 2011). Fourth, we have recently demonstrated that maternal diabetes and high glucose in vitro alter miRNA expression leading to neural stem cell apoptosis (Gu et al., 2015b). However, it is unknown whether altered miRNA expression contributes to ASK1 activation, which is required for high glucose-induced apoptosis.
miR-17 belongs to the miR-17/92 cluster, the members of which are abundantly expressed during embryonic development (Foshay and Gallicano 2009; Mogilyansky and Rigoutsos, 2013). miR-17/92 inhibits cell apoptosis by repressing the expression of pro-apoptotic proteins such as Bim (Li et al., 2014; Ventura et al., 2008). Deletion of the miR-17/92 cluster results in embryonic anomalies that are similar to those observed in diabetic pregnancies (Ventura et al., 2008), suggesting that miR-17 down-regulation may mediate the pro-apoptotic effects of maternal diabetes or high glucose on neural stem cells. It has been shown that miR-20, a member of the miR-17/92 cluster, suppresses ASK1 expression in a disease model of inflammation (Philippe et al., 2013). Because high glucose does not affect ASK1 expression but activates ASK1, we hypothesize that high glucose-induced miR-17 down-regulation induces ASK1 activation.
In this study, we found that maternal diabetes and high glucose in vitro down-regulated miR-17 leading to the up-regulation of its target gene, Thioredoxin-interacting protein (Txnip). Txnip, a thioredoxin (Trx) binding protein, is a negative regulator of the biological function and expression of Trx (Nishiyama et al., 1999). On the other hand, Trx is an inhibitor of ASK1 that is constitutively associated with non-active ASK1 under basal conditions and its dissociation from ASK1 allows subsequent phosphorylation and activation of ASK1 (Saitoh et al., 1998). Here, we revealed a mechanism underlying maternal diabetes- or high glucose-induced ASK1 activation by demonstrating that miR-17 down-regulation-increased Txnip triggers ASK1 activation by suppressing Trx.
MATERIALS AND METHODS
Animals
Wild-type (WT) C57BL/6J mice were purchased from the Jackson Laboratory. Streptozotocin (STZ) from Sigma was dissolved in sterile 0.1 M citrate buffer (pH4.5). The procedures for animal use were approved by the University of Maryland School of Medicine Institutional Animal Care and Use Committee.
Mouse models of diabetic embryopathy
Our mouse model of diabetic embryopathy has been described previously (Yang et al., 2013). Briefly, 10-week old WT female mice were intravenously injected daily with 75 mg/kg STZ over 2 days to induce diabetes. Diabetes was defined as a 12 h fasting blood glucose level of ≥ 14 mM. Male and female mice were paired at 3:00 p.m., and pregnancy was established by the presence of the vaginal plug next morning, and noon of that day was designated as embryonic day 0.5 (E0.5). WT female mice were treated with vehicle injections as non-diabetic controls. On E8.75 (at 6:00 p.m.), mice were euthanized and conceptuses were dissected out of the uteri, embryos with the yolk sacs were removed from the deciduas and then yolk sacs were removed from the embryos. The embryos were used for miRNA profiling using the Exiqon service (www.exiqon.com) as we previously described (Gu et al., 2015b), and RT-qPCR analyses.
Cell culture and treatments
Originally obtained from European Collection of Cell Culture, C17.2 mouse neural stem cells, newborn mouse cerebellar progenitor cells transformed with retroviral v-myc, were maintained in DMEM with 5 mM glucose supplemented with 10% fetal bovine serum, 100 U/ml of penicillin and 100 μg/ml of streptomycin at 37°C in a humidified atmosphere of 5% CO2. Lipofectamine RNAiMAX (Invitrogen) was used according to the manufacturer’s protocol for the transfection of miRNA mimic or inhibitor into cells under 1% fetal bovine serum culture conditions. The mirVana miRNA mimic, the miRNA inhibitor for miR-17 and the negative control (NC) were purchased from Ambion (Life Technologies). Glucose was added to the culture medium at different doses for high glucose conditions. GFP-Txnip plasmid was a gift from Clark Distelhorst (Addgene plasmid no. 18758) (Wang et al., 2006). This plasmid was transfected into cells using Lipofectamine 2000 (Invitrogen).
Plasmid construction
The full-length coding region (CR) or its 3′-UTR and 2 fractions (F1 and F2) in its 3′-UTR with 2 predicted miR-17-binding sites were amplified and subcloned into the pmirGLO Dual-Luciferase miRNA Target Expression Vector (Promega) to generate the pmirGLO-Luc-CR, the pmirGLO-luc-3′-UTR and the pmirGLO-luc-F1/F2. The sequences and orientations of the fragments in the luciferase reporters were confirmed by DNA sequencing and enzyme digestion. Luciferase activity was measured using the Dual-Luciferase Assay System (Promega). Levels of pmirGLO-Luc-CR, 3′-UTR and F1/F2 luciferase activities were normalized to Renilla luciferase activity. All primer sequences for generating these constructs are provided in Supplementary Table S1.
Immunoprecipitation
Cells were lysed in non-denaturing lysis buffer containing 20 mM of Tris-HCl (pH 7.5), 150 mM of NaCl, 1 mM of Na2EDTA, 1 mM of EGTA, 1% Triton, 2.5 mM of sodium pyrophosphate, 1 mM of β−glycerophosphate, 1 mM of Na3VO4, and 1 μg/ml of leupeptin (Cell Signaling technology). Protease inhibitor (Sigma) was added before use. After centrifugation at 12 000 g for 10 min at 4°C, the supernatant was pre-cleaned with protein A/G magnetic beads (Thermo Scientific) for 2 h at 4°C. Five hundred μg of protein extraction were incubated with an antibody to ASK1 (Santa Cruz) or thioredoxin (Abcam) at 4°C overnight. Twenty-five μl of protein A/G magnetic beads was added for immunoprecipitation (IP) at room temperature for 2 h. Precipitated complexes were cleansed in washing buffer (Thermo Scientific), and bound proteins were analyzed by immunoblotting (IB).
Immunoblotting
Equal amounts of protein (30 or 50 μg) from cultured cells or immunoprecipitates were resolved by sodium dodecyl sulfate-polyacryamide gel electrophoresis and transferred onto Immunobilon-P membranes (Millipore). Five μg of Precision Plus Protein Standards (Bio-Rad) was loaded into one lane of the gel. Membranes were incubated in 5% nonfat milk for 1 h and then were incubated for 18 h at 4°C with the following primary antibodies at dilutions of 1:500–1:5000: Txnip (Cell Signaling Technology, 1:1000), p-ASK1 (Santa Cruz, 1:500), ASK1 (Santa Cruz, 1:1000), Trx (Abcam, 1:1000), and caspase 3 (Millipore, 1:1000). Membranes were then exposed to goat anti-rabbit or anti-mouse secondary antibodies. To ensure that equivalent amounts of protein were loaded, membranes were stripped and probed with a mouse antibody against β-actin (Abcam, 1:5000). Signals were detected using the SuperSignal West Femto Maximum Sensitivity Substrate kit (Thermo Scientific). Quantification of blots was performed using VisionWorksLS software (UVP Company). All experiments were repeated in triplicate.
RNA extraction and RT-qPCR
Total RNA was isolated from cells using the Trizol reagent (Ambion) and reverse transcribed using the QuantiTect Reverse Trancription Kit (Qiagen). Reverse transcription for miRNA was performed using the qScript miRNA cDNA Synthesis Kit (Quanta Biosciences). RT-qPCR for Txnip, β-actin, miR-17, and small nuclear RNA U6 was performed using the Maxima SYBR Green/ROX qPCR Master Mix assay (Thermo Scientific). The primers for RT-qPCR are listed in Supplementary Table S2. RT-qPCR and subsequent calculations were performed by a StepOnePlus RT-PCR System (Applied Biosystem).
TUNEL Assay
The TUNEL assay was performed using the In Situ Cell Death Detection Kit (Millipore). Cells were seeded on an 8-well Nunc Lab-Tek Chamber Slide system (Sigma). After transfection and high glucose treatment, cells were fixed with 1% paraformaldehyde in PBS, incubated with TUNEL reagent counterstained with 4′,6-diamidino-2-phenylindole (DAPI) and mounted with aqueous mounting medium (Sigma). TUNEL-positive cells in each well were counted. The percentage of apoptotic cells was calculated as the number of TUNEL-positive (apoptotic) cells divided by the total number of cells in a microscopic field from 3 separate experiments.
Statistics
All experiments were completely randomized designed and repeated in triplicate. Data are presented as means ± SE. One- or two-way ANOVA was performed using the SigmaStat 3.5 software. Most of data were analyzed by 1-way ANOVA. However, 2-way ANOVA was used for Figures 1A, B, D, F, H, J; 2C; and 3B and E. A Tukey test was used to estimate the significance. Statistical significance was indicated when P < 0.05.
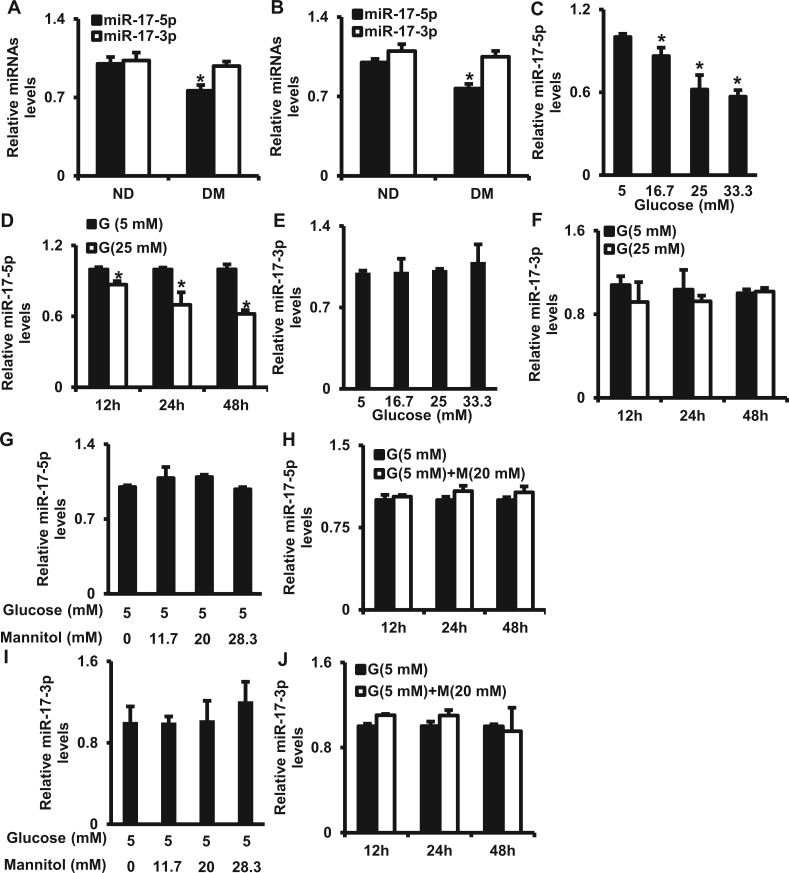
FIG. 1.
Maternal diabetes in vivo and high glucose in vitro down-regulate miR-17. A. miR-17-5p/3p levels in E8.75 embryos determined by the miRNA profiling (n = 3 litters). B. miR-17-5p/3p levels in E8.75 embryos assessed by RT-qPCR. ND, non-diabetic; DM, diabetic mellitus. C. miR-17-5p levels in cells treated with normal glucose (5 mM) or high glucose (16.7, 25, 33.3 mM) for 24 h. D. miR-17-5p levels in cells under normal (5 mM) or high (25 mM) glucose conditions for 12, 24, and 48 h. E. miR-17-3p levels in cells treated with different glucose concentrations for 24 h. F. miR-17-3p levels in cells under normal (5 mM) or high (25 mM) glucose conditions for 12, 24, and 48 h. G. miR-17-5p levels in cells under normal glucose (5 mM) conditions with or without high mannitol (11.7, 20, and 28.3 mM) for 24 h. H. miR-17-5p levels under normal glucose (5 mM) conditions with or without 20 mM of mannitol for 12, 24, and 48 h. I. miR-17-3p levels in cells under normal glucose (5 mM) conditions with or without high mannitol (11.7, 20, and 28.3 mM) for 24 h. J. miR-17-3p levels in cells under normal glucose (5 mM) conditions with or without 20 mM of mannitol for 12, 24, and 48 h. G: glucose, M, mannitol. Experiments were repeated 3 times (n = 3). Values are means ± SEM from 3 separate experiments. *indicates significant difference (P < .05) compared with the ND group or the 5 mM glucose group.
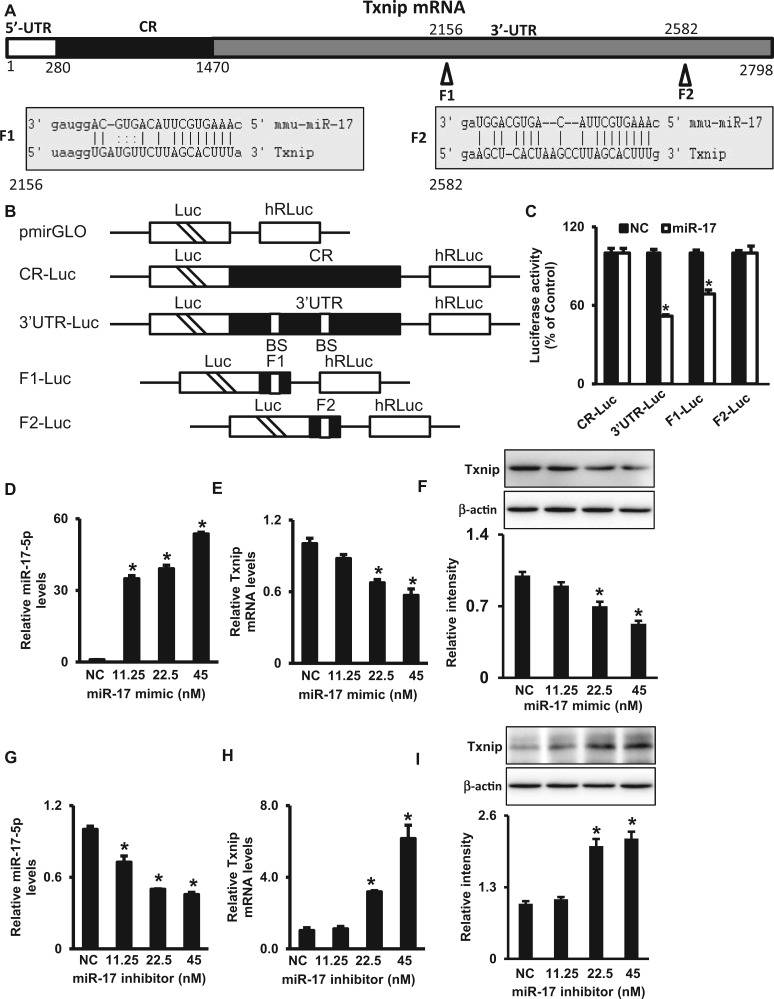
FIG. 2.
miR-17 suppresses Txnip expression. A. Schematic representation of the Txnip mRNA depicting 2-binding sites for miR-17 in its 3′-UTR. CR: coding region; F1 and F2 are 2 miR-17-binding sites. B. Schematic of plasmids of different chimeric firefly luciferase Txnip reporters. BS, miR-17-binding site; CR, coding region; Luc, luciferase. C. Levels of luciferase reporter activities containing Txnip CR, 3′-UTR, F1 or F2. Twenty-four hours after transfection with the miR-17 mimic, cells were transfected with different Txnip luciferase reporter plasmids. Levels of firefly and Renilla luciferase activities were assayed 24 h later. Results were normalized to the Renilla luciferase activity and expressed as means ± SEM from 3 separate experiments. Levels of miR-17 (D), Txnip mRNA (E) and protein (F) after cells were transfected with the miR-17 mimic for 48 h. Levels of miR-17 (G), Txnip mRNA (H) and protein (I) after cells were transfected with the miR-17 inhibitor for 48 h. The quantification of IB is shown by bar graphs. NC, negative control oligos. Experiments were repeated 3 times (n = 3). Values are means ± SE from 3 separate experiments. *indicates significant difference (P < .05) compared with the NC group.
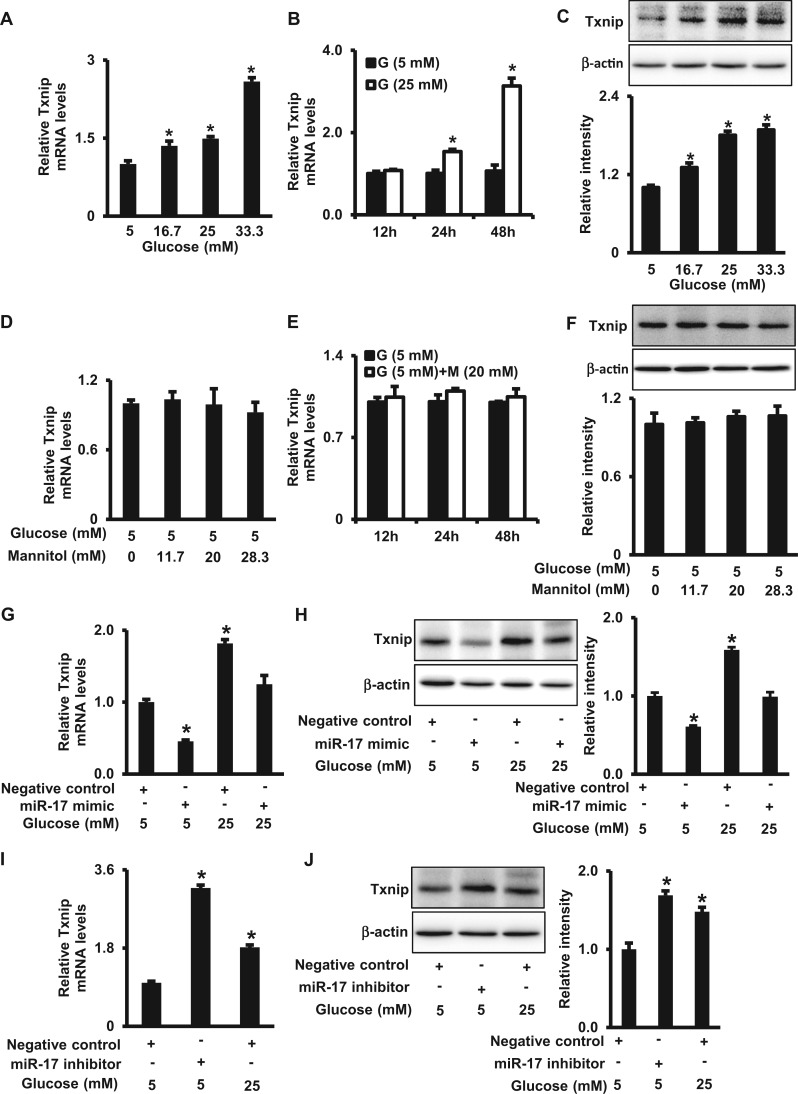
FIG. 3.
High glucose up-regulates Txnip expression by down-regulating miR-17. A. Txnip mRNA levels in cells treated with normal glucose (5 mM) or high glucose (16.7, 25, 33.3 mM) for 24 h. B. Txnip mRNA levels in cells under normal (5 mM) or high (25 mM) glucose conditions for 12, 24, and 48 h. C. Txnip protein levels in cells treated with normal glucose (5 mM) or high glucose (16.7, 25, 33.3 mM) for 48 h. D. Txnip mRNA levels in cells under normal glucose (5 mM) conditions with or without high mannitol (11.7, 20, and 28.3 mM) for 24 h. E. Txnip mRNA levels in cells under normal glucose (5 mM) conditions with or without 20 mM of mannitol for 12, 24, and 48 h. F. Txnip protein levels in cells under normal glucose (5 mM) conditions with or without high mannitol (11.7, 20, and 28.3 mM) for 48 h. Levels of Txnip mRNA (G) and protein (H) in cells treated with different concentrations of glucose (5 or 25 mM) in the absence or presence of the miR-17 mimic for 48 h. Levels of Txnip mRNA (I) and protein (J) in cells treated the miR-17 inhibitor for 48 h. G, glucose; M, mannitol. The quantification of immunoblots is shown by bar graphs. Experiments were repeated 3 times (n = 3). Values are means ± SEM from 3 separate experiments. *indicates significant difference (P < .05) compared with the normal glucose (5 mM) group.
RESULTS
High Glucose Suppresses miR-17 Expression
In our preliminary study, we performed a microarray of global miRNA expression on E8.5 embryos from diabetic and nondiabetic mice. This stage is vital for neurulation during mouse embryonic development. We found that miR-17-5 p but not miR-17-3p was down-regulated in embryos from diabetic dams compared with embryos from nondiabetic dams (Figure 1A). This result was validated by RT-qPCR (Figure 1B).
To test if high glucose in vitro has a similar effect on miR-17-5p expression as that of maternal diabetes, C17.2 neural stem cells were cultured under normal glucose (5 mM) or high glucose (16.7, 25, and 33.3 mM) conditions. High glucose decreased miR-17-5p levels in a dose-dependent manner and the decline of miR-17-5p reached a plateau at 25 mM glucose (Figure 1C). Twenty-five mM glucose is comparable to the high blood glucose level (average: 26 mM of glucose) of diabetic dams. A time-course study on the effect of 25 mM glucose showed that miR-17-5p was down-regulated at 12, 24, and 48 h (Figure 1D). We did not find any changes in miR-17-3p levels under high glucose conditions (Figs. 1E and F). In addition, we used mannitol as an osmotic control for glucose. High mannitol had no effect on the expression of miR-17-5p and miR-17-3p levels (Figs. 1G–J).
A precursor miRNA produces a mature miRNA (a guide strand for gene regulation) and a passenger strand, which is degraded and does not play a role in gene regulation. According to the miRNA database (www.mirbase.org), miR-17-5p is the mature miR-17 and miR-17-3p is the passenger strand. Therefore, we subsequently used miR-17 instead of miR-17-5p.
Txnip Is a Target Gene of miR-17
Bioinformatic target prediction algorithm (miRanda, www.microRNA.org) reveals that Txnip is a predicted target gene of miR-17. There are 2 potential-binding sites of miR-17 in the 3′-UTR of Txnip (Figure 2A). To test if Txnip is a true target of miR-17, we used luciferase reporter constructs to investigate if miR-17 can directly regulate Txnip expression. miRNAs are able to repress gene expression by binding to seed site sequences located within the 3'-UTRs of mRNAs. Fractions of the CR and 3′-UTR of Txnip mRNA or the specific binding sites (Fraction 1 [F1] and F2) of miR-17 were subcloned into the pmirGLO dual-luciferase miRNA target expression vector to generate CR-luc, 3′-UTR-luc, F1-luc and F2-luc reporter constructs as depicted in Figure 2B. The miR-17 mimic and the luciferase constructs were co-transfected into cells. The miR-17 mimic significantly decreased the luciferase activities of 3′-UTR-luc and F1-luc reporters but failed to inhibit the activities of CR-luc and F2-luc reporters (Figure 2C). This indicates that miR-17 repressed Txnip expression by interacting with the F1 binding site in the Txnip 3′-UTR.
The repression of Txnip expression by miR-17 was further verified by the transfection with the miR-17 mimic and inhibitor. miR-17 levels increased markedly from the transfection with the miR-17 mimic (Figure 2D). Txnip mRNA and protein levels were significantly decreased by the miR-17 mimic (Figs. 2E and F). On the other hand, miR-17 levels were decreased by transfection with the miR-17 inhibitor (Figure 2G), and Txnip mRNA and protein levels increased accordingly (Figs. 2H and I). Altogether, these results indicate that miR-17 represses Txnip expression through its interaction with 1 specific binding site of the Txnip 3′-UTR and subsequent degradation of mRNA.
High Glucose Increases Txnip Expression Through miR-17
Since high glucose down-regulates miR-17, we sought to investigate if high glucose also regulates the miR-17 target gene Txnip expression. Cells were treated with normal (5 mM) and high (16.7, 25, and 33.3 mM) glucose for 48 h. High glucose increased Txnip mRNA and protein levels in a dose-dependent manner (Figs. 3A and C). A time-course study of the effect of 25 mM glucose showed that Txnip mRNA was up-regulated at 24 and 48 h but not 12 h (Figure 3B). In contrast, mannitol as an osmotic control for glucose did not affect Txnip mRNA and protein levels (Figs. 3D–F).
To explore if miR-17 down-regulation mediates the stimulative effect of high glucose on Txnip expression, we restored miR-17 expression by transfecting cells with the miR-17 mimic under normal (5 mM) or high (25 mM) glucose conditions. The miR-17 mimic suppressed high glucose-induced increase of Txnip in mRNA and protein levels (Figs. 3G and H). Conversely, the miR-17 inhibitor mimicked the stimulative effect of high glucose on Txnip expression (Figs. 3I and J). Thus, these findings support that high glucose induces Txnip expression through repression of miR-17.
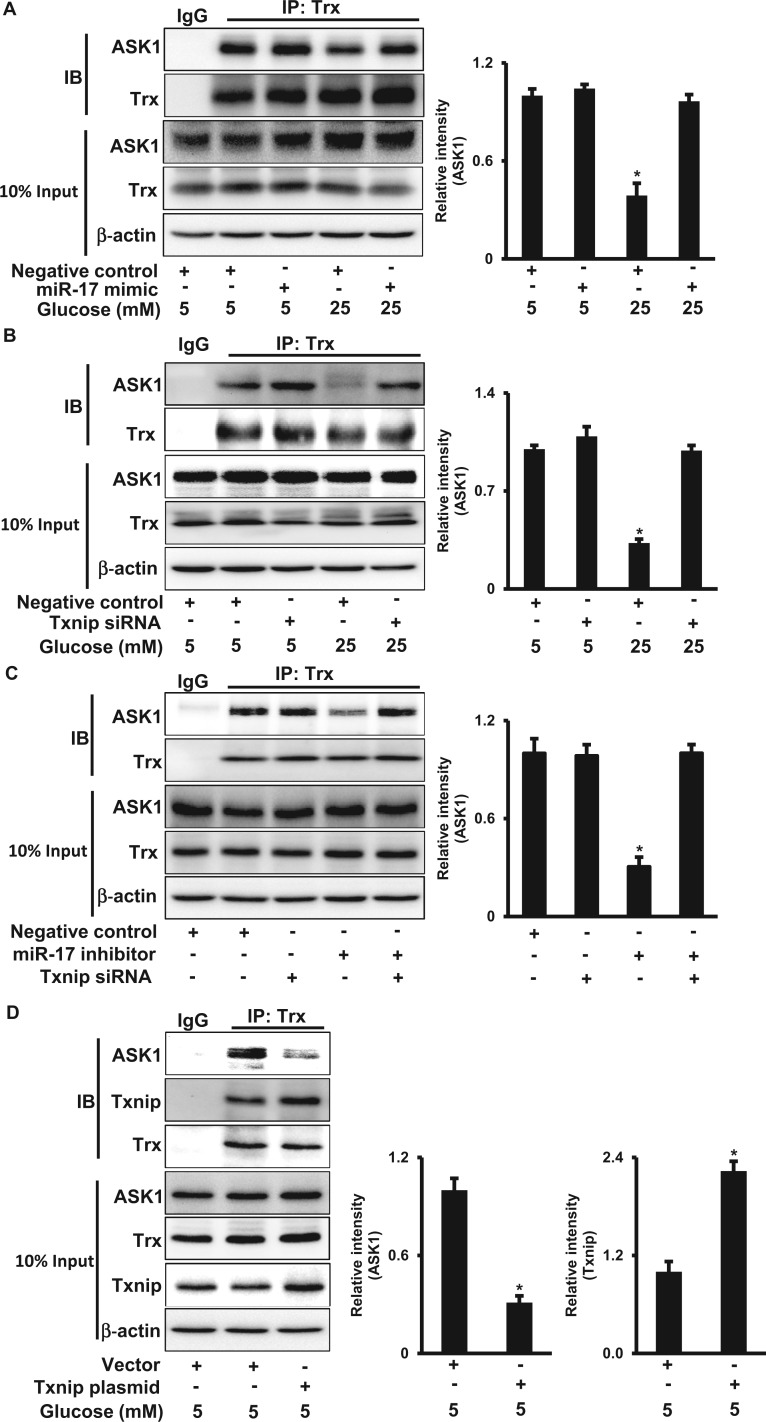
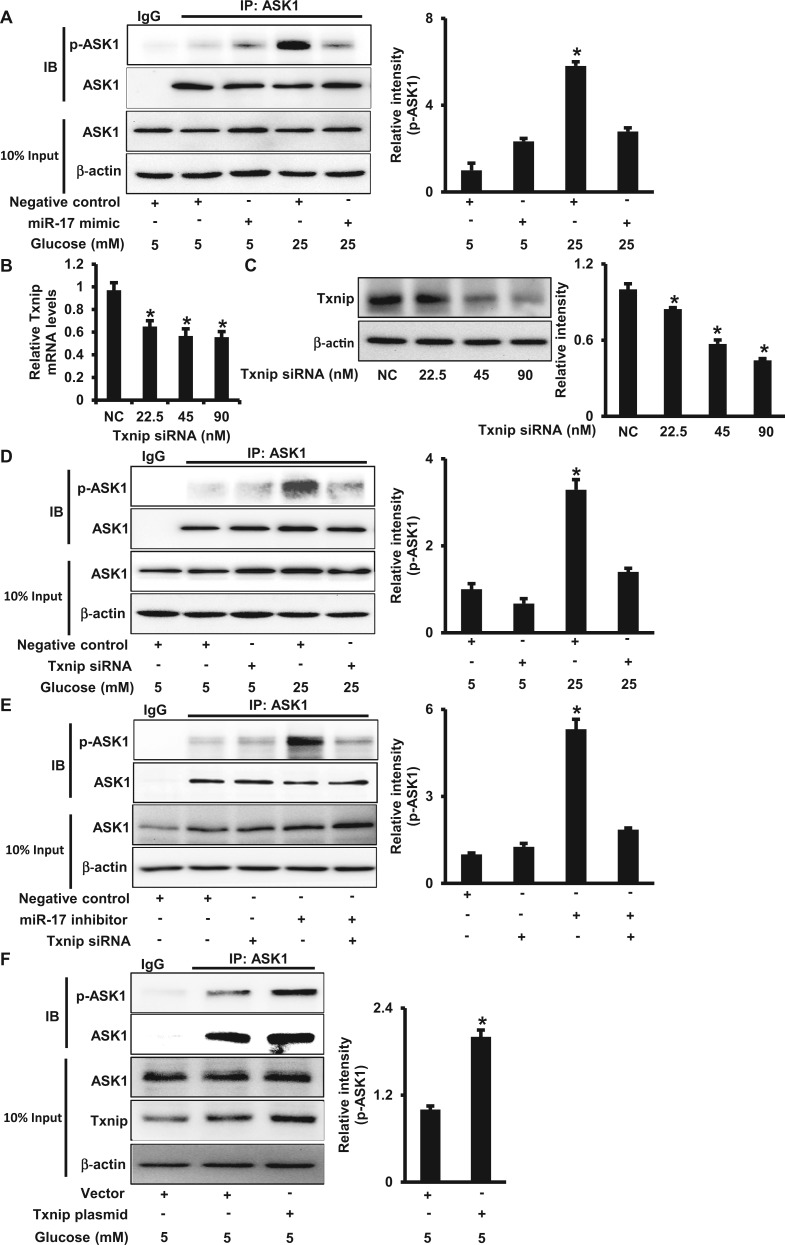
Txnip Sequesters Trx From the Trx-ASK1 Complex Leading to ASK1 Activation
To elucidate how high glucose activates ASK1, we assessed the interaction between ASK1 and Trx, an inhibitor of ASK1 that is constitutively associated with nonactive ASK1 under basal conditions and whose dissociation from ASK1 allows subsequent phophorylation and activation of ASK1 (Saitoh et al., 1998). The amount of ASK1 in Trx immunoprecipitates from high glucose (25 mM)-treated cell lysates was significantly lower than that in normal glucose (5 mM)-treated cell lysates (Figure 4A). The decreased association between Trx and ASK1 was not due to reduced abundances of endogenous ASK1 and Trx because high glucose had no effect on total ASK1 and Trx protein levels (Figure 4A).
FIG. 4.
Txnip sequesters Thioredoxin from ASK1 under high glucose conditions. A. ASK1 protein abundance in Trx immunoprecipitates from cells treated with different doses of glucose (5 or 25 mM) in the absence or presence of the miR-17 mimic (A), or Txnip siRNA (B) for 48 h. C. ASK1 protein abundance in Trx immunoprecipitates from cells transfected with the miR-17 inhibitor in the absence or presence of Txnip siRNA for 48 h. D. ASK1 and Txnip protein abundances in cells transfected with the Txnip plasmid under normal glucose (5 mM) conditions for 48 h. IP was performed using the anti-Trx antibody. Quantification of IB is shown in the bar graphs. Experiments were repeated 3 times (n = 3). Values are means ± SE from 3 separate experiments. *indicates significant difference (P < .05) compared with the other group(s).
Next, we tested whether miR-17 and Txnip are responsible for the dissociation between Trx and ASK1 induced by high glucose. Both the miR-17 mimic and Txnip siRNA knockdown restored the binding between Trx and ASK1 under high glucose conditions (Figs. 4A and B). On the other hand, the miR-17 inhibitor treatment led to the dissociation between Trx and ASK1, and the effect was suppressed by Txnip siRNA knockdown (Figure 4C). To uncover whether high glucose-increased Txnip through miR-17 suppression mediates the dissociation between Trx and ASK1, Txnip was overexpressed in cells. Txnip overexpression increased the binding between Txnip and Trx, whereas the interaction between Trx and ASK1 was significantly decreased (Figure 4D). Therefore, our data suggest that the increase of Txnip protein levels by high glucose and miR-17 sequesters Trx from ASK1 leading to ASK1 activation.
High Glucose Activates ASK1 via the miR-17-Txnip Circuit
Our previous studies have demonstrated that maternal diabetes activates ASK1 leading to apoptosis in the developing neuroepithelium (Yang et al., 2013). To investigate whether miR-17 and Txnip contribute to maternal diabetes-induced ASK1 activation, which is responsible for high glucose-induced apoptosis, the phosphorylation of ASK1 was assessed. To detect the activation of ASK1 with greater easily, we enriched total ASK1 protein through IP and then determined phosphorylation at the Thr845 of ASK1, which is essential for the catalytic activity of ASK1 and the induction of apoptosis (Tobiume et al., 2002). High glucose induced ASK1 phorphorylation under treatment with high glucose (25 mM), and the effect was inhibited by the miR-17 mimic (Figure 5A).
FIG. 5.
High glucose activates ASK1 through the miR-17-Txnip circuit. A. P-ASK1 protein abundance in cells treated with different doses of glucose (5 or 25 mM) in the absence or presence of the miR-17 mimic for 48 h. IP was performed using the anti-ASK1 antibody. Changes in p-ASK1 were determined by IB. Levels of Txnip mRNA (B) and protein (C) in cells transfected with Txnip siRNA for 48 h. P-ASK1 protein abundance in cells treated with different doses of glucose (5 or 25 mM) in the absence or presence of Txnip siRNA (D), or with co-transfection of the miR-17 inhibitor (E). F. P-ASK1 protein abundance in cells transfected with the Txnip plasmid under normal glucose (5 mM) conditions for 48 h. The quantification of immunoblots is shown in the bar graphs. NC, negative control oligos. Experiments were repeated 3 times (n = 3). Values are means ± SEM from 3 separate experiments. *indicates significant difference (P < .05) compared with the NC group or the other group(s).
Because Txnip is a target gene of miR-17 and high glucose increased Txnip expression, we tested if suppressing Txnip expression inhibits high glucose-induced ASK1 activation. Ninety nM of Txnip siRNA treatment reduced Txnip mRNA and protein levels to approximately 50% of those in the control groups (Figs. 5B and C). When Txnip was silenced, high glucose-induced ASK1 phosphorylation was suppressed (Figure 5D).
Additionally, the reduction of miR-17 by the miR-17 inhibitor promoted ASK1 phosphorylation, and the effect was blocked by the Txnip siRNA (Figure 5E). Txnip overexpression by transfecting the Txnip plasmid also induced phorphorylation of ASK1 (Figure 5F). Taken together, these data indicate that high glucose induces ASK1 activation through down-regulation of miR-17 and consequent up-regulation of Txnip.
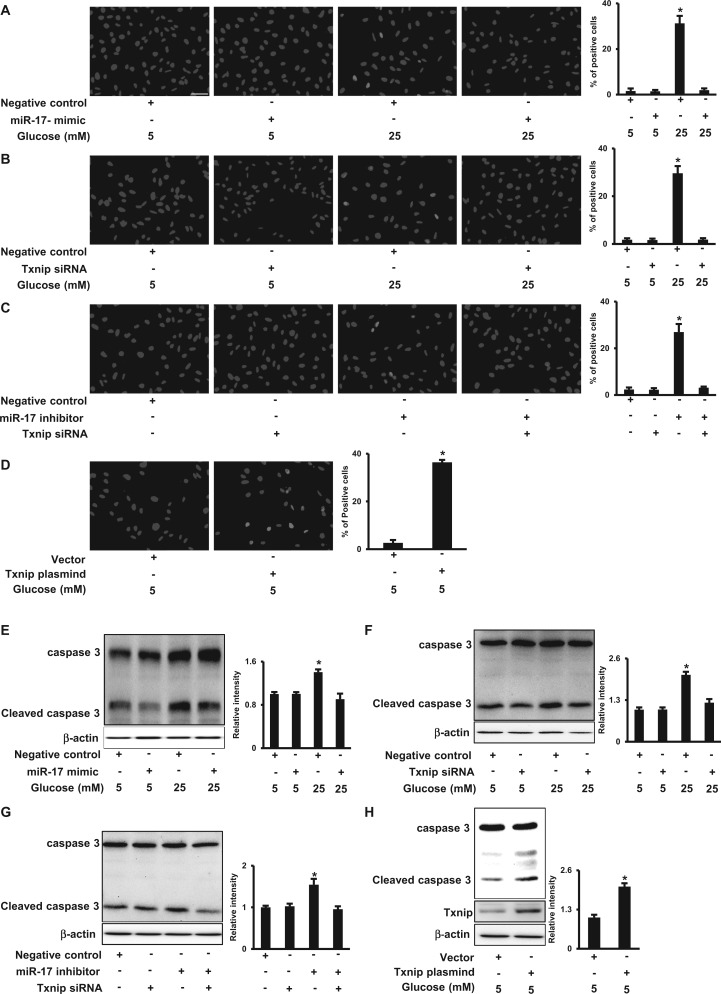
The miR-17-Txnip Circuit Mediates the Pro-apoptotic Effect of High Glucose
To define the biological effect of high glucose-mediated decrease of miR-17, increase of Txnip and subsequent ASK1 activation, we assessed the number of apoptotic cells and caspase cleavage. When cells were exposed to high glucose, apoptotic cells were robustly present, whereas the miR-17 mimic protected cells from high glucose-induced aopotosis (Figure 6A). Likewise, Txnip siRNA knockdown significantly reduced the high glucose-increased apoptotic cell number (Figure 6B). Moreover, the miR-17 inhibitor triggered cell apoptosis, and the effect was attenuated by Txnip siRNA knockdown (Figure 6C). Txnip overexpression also induced cell apoptosis in normal glucose (5 mM) conditions (Figure 6D). High glucose increased the abundance of cleaved caspase 3, and both the miR-17 mimic and Txnip siRNA knockdown suppressed high glucose-induced caspase 3 cleavage (Figs. 6E and F). Txnip siRNA knockdown prevented miR-17 inhibitor-increased caspase 3 cleavage (Figure 6G). Meanwhile, Txnip overexpression induced caspase 3 cleavage (Figure 6H). These data suggest that high glucose induces apoptosis through the miR-17-Txnip-ASK1 pathway.
FIG. 6.
High glucose induces apoptosis by down-regulating miR-17 and consequently up-regulating Txnip. A–D. Representative images and quantifications of TUNEL positive cells. Cells treated with different doses of glucose (5 or 25 mM) in the absence or presence of the miR-17 mimic (A), or in the absence or presence of Txnip siRNA (B). In C, cells were transfected with the miR-17 inhibitor in the absence or presence of Txnip siRNA. In D, cells were transfected with the Txnip plasmid under normal glucose (5 mM) conditions for 48 h. DAPI nuclei staining. Bar, 20 µm. E–H. Levels of cleaved caspase 3. Cells were treated with different doses of glucose (5 or 25 mM) in the absence or presence of the miR-17 mimic (E), or in the absence or presence of Txnip siRNA (F). In G, cells were transfected with the miR-17 inhibitor in the absence or presence of Txnip siRNA, and in H, cells were transfected with the Txnip plasmid under normal glucose (5 mM) conditions. Quantification of TUNEL positive cells or IB is shown in the bar graphs. Experiments were repeated 3 times (n = 3). Values are means ± SE from 3 separate experiments. *indicates significant difference (P < .05) compared with the other group(s).
DISCUSSION
Maternal diabetes-induced malformations may involve more than 1 organ and frequently result in significant disability or death in offspring (Eriksson et al., 2000; Ylinen et al., 1984). Because the neural folds and the heart develop early during embryogenesis, a higher incidence of malformations is observed in these organs (Eriksson et al., 2000; Ylinen et al., 1984). Multiple studies including ours have revealed that excessive cell apoptosis in the developing neuroepithelium contributes to the abnormal development of structures in embryos of diabetic animals (Eriksson et al., 2000; Fine et al., 1999; Forsberg et al., 1998; Li et al., 2012, 2013; Moley, 2001; Sun et al., 2005; Yang et al., 2013). These findings strongly support the hypothesis that high glucose causes damage to neuroepithelial cells or neural stem cells, leading to apoptosis and, ultimately, NTDs. Our previous studies have demonstrated that high glucose of diabetes induces apoptosis in neuroepithelial cells through the ASK1-JNK1/2-FoxO3a-TRADD-caspase 8 pathway (Li et al., 2012, 2013; Yang et al., 2013). It is suggested that ASK1 activation is an initial event for apoptosis. In this study, we demonstrated that ASK1 activation was associated with high glucose-suppressed miR-17 and -increased Txnip. Txnip is up-regulated by high glucose through miR-17 down-regulation. Increased Txnip is bound to Trx, which leads to the dissociation of Trx from ASK1 leading to ASK1 activation. Thus, high glucose induces ASK1 activation via the miR-17-Txnip-Trx pathway.
ASK1 belongs to the family of MAPK kinase kinase kinase (MAP3K) (Widmann et al., 1999). ASK1-mediated apoptosis is involved in the pathogenesis of several diseases such as brain ischemia (Zhang et al., 2003), ischemic heart disease (Watanabe et al., 2005) and Alzheimer’s disease (Kadowaki et al., 2005). Most recently it has been shown that high glucose-activated ASK1 mediates endothelial cell senescence (Yokoi et al., 2006). These findings are consistent with our findings that ASK1 functions as a mediator of diabetes-induced embryonic malformations. ASK1 can be activated through different mechanisms. One mechanism is that oxidative stress directly induces phosphorylation of Thr845 in the activation loop of ASK1, a process correlated with enhanced ASK1 activity and increased apoptosis (Tobiume et al., 2002). Another is that Trx inhibits ASK1 through a direct interaction, and the interaction between Trx and ASK1 depends on the redox status of Trx (Saitoh et al., 1998). ROS oxidizes the cysteine residues of Trx, which induce the dissociation of Trx from ASK1, thereby leading to ASK1 activation (Saitoh et al., 1998). We uncovered a new mechanism of ASK1 activation in which the dissociation of Trx from ASK1 is induced by increased Txnip, a Txnip (Nishiyama et al., 1999), leading to ASK1 phosphorylation and subsequent apoptosis in neural stem cells under high glucose conditions.
Txnip was first identified as a 1,25-dihydroxyvitamin D-3 inducible gene (Chen and DeLuca, 1994). Using the yeast 2-hybrid system, Txnip was revealed as a Trx-binding protein (Nishiyama et al., 1999). Txnip is one of the genes that are most highly inducible by diabetes and high glucose in various tissues (Chen et al., 2008; Cheng et al., 2006; Kobayashi et al., 2003; Price et al., 2006; Shalev et al., 2002; Singh, 2013). The induction of Txnip by high glucose or diabetes is carried out through several pathways (Singh, 2013). These pathways focus on the Txnip promoter, which contains various transcription factor binding sites, such as E-Box (also known as carbohydrate response element), Foxo element, and anti-oxidant response element (Singh, 2013). Txnip transcription induced by high glucose involves an array of transcription factors (Singh, 2013). Our study elucidates that Txnip up-regulation by high glucose is through a post-transcriptional mechanism, in which high glucose-induced decay of miR-17 contributes to the increase of Txnip. miR-17 represses Txnip expression by a direct interaction with one specific binding site in the Txnip 3′-UTR. This interaction leads to subsequent degradation of Txnip mRNA.
The physiological function of Txnip is not yet fully understood. However, Txnip has emerged as a new pro-apoptotic factor, especially for diabetes-mediated pancreatic β-cell apoptosis (Chen et al., 2008, 2010; Harada et al., 2015; Lerner et al., 2012; Minn et al., 2005; Shalev, 2008; Wang et al., 2006). Txnip is implicated in a critical link between glucose toxicity and pancreatic β-cell apoptosis (Chen et al., 2008). Lack of Txnip protects pancreatic β-cells against glucotoxicity (Chen et al., 2010; Masson et al., 2009; Shalev, 2008). High glucose leads to a significant elevation of Txnip and apoptosis in pancreatic islets in vivo and pancreatic β-cells in vitro (Chen et al., 2008, 2010; Shalev, 2008). The major mechanism underlying Txnip-induced apoptosis is via the mitochondrial pathway but not the ER stress pathway because Txnip overexpression induces mitochondrial cytochrome c release into cytosolic fractions, whereas there is no change in ER stress marker expression (Chen et al., 2008). Another indication is that the lack of Txnip protects β-cell apoptosis induced by staurosporine, a well-known stimulus of the mitochondrial death pathway, but not by thapsigargin, an ER stress inducer (Chen et al., 2010). In this study, we identified Txnip as a critical factor mediator of high glucose-induced neural stem cell apoptosis through ASK1 activation. We have previously shown that ASK1 activation induces apoptosis through 2 different pathways in neuroepithelial cells or neural stem cells (Wang et al., 2015b; Yang et al., 2015). One is that activated ASK1 phosphorylates c-Jun N-terminal kinase 1/2 and induces translocation of pro-apoptotic Bcl-2 family members to the mitochondria, resulting in apoptosis (Wang et al., 2015b; Yang et al., 2015). Another is that activated ASK1 increases the nuclear translocation of FoxO3a, which induces TRADD expression. Up-regulation of TRADD leads to caspase 8 cleavage and apoptosis (Wang et al., 2015b; Yang et al., 2015). Therefore, our study suggests that Txnip induces apoptosis by the mitochondrial pathway and the FoxO3a-TRADD pathway, both of which are activated by ASK1 in neural stem cells.
Members of the miR-17/92 cluster include miR-17, miR-18a, miR-19a, miR-20a, miR-19b-1, and miR-92a-1 (Concepcion et al., 2012; Mendell, 2008; Mogilyansky and Rigoutsos, 2013). The miR-17/92 cluster was initially linked to cancer pathogenesis (Mogilyansky and Rigoutsos, 2013), and its role in embryonic development is beginning to emerge (Mendell, 2008). The miR-17/92 cluster is highly expressed in embryonic cells (Ventura et al., 2008). Loss-of-function of the miR-17/92 cluster results in smaller embryos and mediates postnatal death (de Pontual et al., 2011; Ventura et al., 2008). This is likely due to severe developmental defects and widespread apoptosis (de Pontual et al., 2011; Ventura et al., 2008). These observations are consistent with our present finding that high glucose-suppressed miR-17 triggers cell apoptosis. miR-17 is reduced in neurulation stage embryos exposed to maternal diabetes. Because apoptosis is the central mechanism of maternal diabetes-induced NTDs (Gabbay-Benziv et al., 2015; Wang et al., 2015b; Yang et al., 2015), it is highly possible that miR-17 down-regulation is critically involved in the induction of NTDs in diabetic pregnancies.
Our previous studies have demonstrated that ER stress plays a critical role in the etiology of maternal diabetes-induced structural birth defects (Gu et al., 2015a; Li et al., 2013; Wang et al., 2015d, 2015e,f). Maternal diabetes in vivo or high glucose in vitro induces ER stress by activating the unfolded protein response pathways including the inositol-requiring enzyme-1α (IRE1α)-initiated pathway in the developing embryo (Gu et al., 2015a; Zhong et al., 2015). IRE1α activation triggers its endoribonuclease activity, which causes a rapid decay of a group of miRNAs including miR-17, miR-34a, and miR-125b (Upton et al., 2012). Therefore, IRE1α may be responsible for high glucose-induced miR-17 down-regulation.
In summary, our study revealed that high glucose activated ASK1 by suppressing miR-17 expression and increasing the expression of the miR-17 target gene Txnip. Txnip was competently bound to the endogenous ASK1 inhibitor Trx leading to the dissociation between ASK1 and Trx. The events collectively led to ASK1 activation. Our findings elucidate a unique mechanism underlying high glucose-induced ASK1 activation and cell apoptosis, in which the miR-17-Txnip circuit induces ASK1 activation.
SUPPLEMENTARY DATA
Supplementary data are available online at http://toxsci.oxfordjournals.org/.
FUNDING
This work was supported by NIH R01DK083243, R01DK101972, R01DK103024, and a Basic Science Award (1-13-BS-220), American Diabetes Association.
Supplementary Material
REFERENCES
- Bartel D. P. (2009). MicroRNAs: Target recognition and regulatory functions. Cell 136, 215–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., Fontes G., Saxena G., Poitout V., Shalev A. (2010). Lack of TXNIP protects against mitochondria-mediated apoptosis but not against fatty acid-induced ER stress-mediated beta-cell death. Diabetes 59, 440–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., Saxena G., Mungrue I. N., Lusis A. J., Shalev A. (2008). Thioredoxin-interacting protein: a critical link between glucose toxicity and beta-cell apoptosis. Diabetes 57, 938–944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen K. S., DeLuca H. F. (1994). Isolation and characterization of a novel cDNA from HL-60 cells treated with 1,25-dihydroxyvitamin D-3. Biochim. Biophys. Acta 1219, 26–32. [DOI] [PubMed] [Google Scholar]
- Cheng D. W., Jiang Y., Shalev A., Kowluru R., Crook E. D., Singh L. P. (2006). An analysis of high glucose and glucosamine-induced gene expression and oxidative stress in renal mesangial cells. Arch. Physiol. Biochem. 112, 189–218. [DOI] [PubMed] [Google Scholar]
- Concepcion C. P., Bonetti C., Ventura A. (2012). The microRNA-17-92 family of microRNA clusters in development and disease. Cancer J. 18, 262–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Correa A., Gilboa S. M., Besser L. M., Botto L. D., Moore C. A., Hobbs C. A., Cleves M. A., Riehle-Colarusso T. J., Waller D. K., Reece E. A. (2008). Diabetes mellitus and birth defects. Am. J. Obstet. Gynecol. 199, 237 e1–237 e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Pontual L., Yao E., Callier P., Faivre L., Drouin V., Cariou S., Van Haeringen A., Genevieve D., Goldenberg A., Oufadem M., et al. (2011). Germline deletion of the miR-17 approximately 92 cluster causes skeletal and growth defects in humans. Nat. Genet. 43, 1026–1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eriksson U. J., Borg L. A., Cederberg J., Nordstrand H., Siman C. M., Wentzel C., Wentzel P. (2000). Pathogenesis of diabetes-induced congenital malformations. Upsala J. Med. Sci. 105, 53–84. [DOI] [PubMed] [Google Scholar]
- Fine E. L., Horal M., Chang T. I., Fortin G., Loeken M. R. (1999). Evidence that elevated glucose causes altered gene expression, apoptosis, and neural tube defects in a mouse model of diabetic pregnancy. Diabetes 48, 2454–2462. [DOI] [PubMed] [Google Scholar]
- Forsberg H., Eriksson U. J., Welsh N. (1998). Apoptosis in embryos of diabetic rats. Pharmacol. Toxicol. 83, 104–111. [DOI] [PubMed] [Google Scholar]
- Foshay K. M., Gallicano G. I. (2009). miR-17 family miRNAs are expressed during early mammalian development and regulate stem cell differentiation. Dev. Biol. 326, 431–443. [DOI] [PubMed] [Google Scholar]
- Gabbay-Benziv R., Reece E. A., Wang F., Yang P. (2015). Birth defects in pregestational diabetes: Defect range, glycemic threshold and pathogenesis. World J. Diabetes 6, 481–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gareskog M., Cederberg J., Eriksson U. J., Wentzel P. (2007). Maternal diabetes in vivo and high glucose concentration in vitro increases apoptosis in rat embryos. Reprod. Toxicol. 23, 63–74. [DOI] [PubMed] [Google Scholar]
- Gu H., Li H., Zhang L., Luan H., Huang T., Wang L., Fan Y., Zhang Y., Liu X., Wang W., Yuan Z. (2012). Diagnostic role of microRNA expression profile in the serum of pregnant women with fetuses with neural tube defects. J. Neurochem. 122, 641–649. [DOI] [PubMed] [Google Scholar]
- Gu H., Yu J., Dong D., Zhou Q., Wang J. Y., Fang S., Yang P. (2015a). High glucose-repressed CITED2 expression through miR-200b triggers the unfolded protein response and endoplasmic reticulum stress. Diabetes pii, db15-0108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu H., Yu J., Dong D., Zhou Q., Wang J. Y., Yang P. (2015b). The miR-322-TRAF3 circuit mediates the pro-apoptotic effect of high glucose on neural stem cells. Toxicol. Sci. 144, 186–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harada N., Katsuki T., Takahashi Y., Masuda T., Yoshinaga M., Adachi T., Izawa T., Kuwamura M., Nakano Y., Yamaji R., Inui H. (2015). Androgen receptor silences thioredoxin-interacting protein and competitively inhibits glucocorticoid receptor-mediated apoptosis in pancreatic beta-Cells. J. cell. Biochem. 116, 998–1006. [DOI] [PubMed] [Google Scholar]
- Kadowaki H., Nishitoh H., Urano F., Sadamitsu C., Matsuzawa A., Takeda K., Masutani H., Yodoi J., Urano Y., Nagano T., Ichijo H. (2005). Amyloid beta induces neuronal cell death through ROS-mediated ASK1 activation. Cell Death Differ. 12, 19–24. [DOI] [PubMed] [Google Scholar]
- Kobayashi T., Uehara S., Ikeda T., Itadani H., Kotani H. (2003). Vitamin D3 up-regulated protein-1 regulates collagen expression in mesangial cells. Kidney Int. 64, 1632–1642. [DOI] [PubMed] [Google Scholar]
- Lee R. C., Feinbaum R. L., Ambros V. (1993). The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 75, 843–854. [DOI] [PubMed] [Google Scholar]
- Lerner A. G., Upton J. P., Praveen P. V., Ghosh R., Nakagawa Y., Igbaria A., Shen S., Nguyen V., Backes B. J., Heiman M., et al. (2012). IRE1alpha induces thioredoxin-interacting protein to activate the NLRP3 inflammasome and promote programmed cell death under irremediable ER stress. Cell Metab. 16, 250–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R., Chase M., Jung S. K., Smith P. J., Loeken M. R. (2005). Hypoxic stress in diabetic pregnancy contributes to impaired embryo gene expression and defective development by inducing oxidative stress. Am. J. Physiol. Endocrinol. Metab. 289, E591–E599. [DOI] [PubMed] [Google Scholar]
- Li X., Weng H., Xu C., Reece E. A., Yang P. (2012). Oxidative stress-induced JNK1/2 activation triggers proapoptotic signaling and apoptosis that leads to diabetic embryopathy. Diabetes 61, 2084–2092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Xu C., Yang P. (2013). c-Jun NH2-terminal kinase 1/2 and endoplasmic reticulum stress as interdependent and reciprocal causation in diabetic embryopathy. Diabetes 62, 599–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Choi P. S., Casey S. C., Dill D. L., Felsher D. W. (2014). MYC through miR-17-92 suppresses specific target genes to maintain survival, autonomous proliferation, and a neoplastic state. Cancer Cell 26, 262–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magenta A., Cencioni C., Fasanaro P., Zaccagnini G., Greco S., Sarra-Ferraris G., Antonini A., Martelli F., Capogrossi M. C. (2011). miR-200c is upregulated by oxidative stress and induces endothelial cell apoptosis and senescence via ZEB1 inhibition. Cell Death Differ. 18, 1628–1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masson E., Koren S., Razik F., Goldberg H., Kwan E. P., Sheu L., Gaisano H. Y., Fantus I. G. (2009). High beta-cell mass prevents streptozotocin-induced diabetes in thioredoxin-interacting protein-deficient mice. Am. J. Physiol. Endocrinol. Metab. 296, E1251–E1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendell J. T. (2008). miRiad roles for the miR-17-92 cluster in development and disease. Cell 133, 217–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minn A. H., Hafele C., Shalev A. (2005). Thioredoxin-interacting protein is stimulated by glucose through a carbohydrate response element and induces beta-cell aFpoptosis. Endocrinology 146, 2397–2405. [DOI] [PubMed] [Google Scholar]
- Mogilyansky E., Rigoutsos I. (2013). The miR-17/92 cluster: a comprehensive update on its genomics, genetics, functions and increasingly important and numerous roles in health and disease. Cell Death Differ. 20, 1603–1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moley K. H. (2001). Hyperglycemia and apoptosis: mechanisms for congenital malformations and pregnancy loss in diabetic women. Trends Endocrinol Metab. 12, 78–82. [DOI] [PubMed] [Google Scholar]
- Mukhopadhyay P., Brock G., Appana S., Webb C., Greene R. M., Pisano M. M. (2011). MicroRNA gene expression signatures in the developing neural tube. Birth Defects Res. A Clin. Mol. Teratol. 91, 744–762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiyama A., Matsui M., Iwata S., Hirota K., Masutani H., Nakamura H., Takagi Y., Sono H., Gon Y., Yodoi J. (1999). Identification of thioredoxin-binding protein-2/vitamin D(3) up-regulated protein 1 as a negative regulator of thioredoxin function and expression. J. Biol. Chem. 274, 21645–21650. [DOI] [PubMed] [Google Scholar]
- Pavlinkova G., Salbaum J. M., Kappen C. (2009). Maternal diabetes alters transcriptional programs in the developing embryo. BMC Genomics 10, 274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philippe L., Alsaleh G., Pichot A., Ostermann E., Zuber G., Frisch B., Sibilia J., Pfeffer S., Bahram S., Wachsmann D., et al. (2013). MiR-20a regulates ASK1 expression and TLR4-dependent cytokine release in rheumatoid fibroblast-like synoviocytes. Ann. Rheum. Dis. 72, 1071–1079. [DOI] [PubMed] [Google Scholar]
- Price S. A., Gardiner N. J., Duran-Jimenez B., Zeef L. A., Obrosova I. G., Tomlinson D. R. (2006). Thioredoxin interacting protein is increased in sensory neurons in experimental diabetes. Brain Res. 1116, 206–214. [DOI] [PubMed] [Google Scholar]
- Saitoh M., Nishitoh H., Fujii M., Takeda K., Tobiume K., Sawada Y., Kawabata M., Miyazono K., Ichijo H. (1998). Mammalian thioredoxin is a direct inhibitor of apoptosis signal-regulating kinase (ASK) 1. EMBO J. 17, 2596–2606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salbaum J. M., Kappen C. (2010). Neural tube defect genes and maternal diabetes during pregnancy. Birth Defects Res. A Clin. Mol. Teratol. 88, 601–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalev A. (2008). Lack of TXNIP protects beta-cells against glucotoxicity. Biochem. Soc. Trans. 36(Pt 5), 963–965. [DOI] [PubMed] [Google Scholar]
- Shalev A., Pise-Masison C. A., Radonovich M., Hoffmann S. C., Hirshberg B., Brady J. N., Harlan D. M. (2002). Oligonucleotide microarray analysis of intact human pancreatic islets: Identification of glucose-responsive genes and a highly regulated TGFbeta signaling pathway. Endocrinology 143, 3695–3698. [DOI] [PubMed] [Google Scholar]
- Singh L. P. (2013). Thioredoxin interacting protein (TXNIP) and pathogenesis of diabetic retinopathy. J. Clin. Exp. Ophthalmol. 4, 287 doi:10.4172/2155-9570.1000287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun F., Kawasaki E., Akazawa S., Hishikawa Y., Sugahara K., Kamihira S., Koji T., Eguchi K. (2005). Apoptosis and its pathway in early post-implantation embryos of diabetic rats. Diabetes Res. Clin. Pract. 67, 110–118. [DOI] [PubMed] [Google Scholar]
- Tobiume K., Saitoh M., Ichijo H. (2002). Activation of apoptosis signal-regulating kinase 1 by the stress-induced activating phosphorylation of pre-formed oligomer. J. Cell. Physiol. 191, 95–104. [DOI] [PubMed] [Google Scholar]
- Upton J. P., Wang L., Han D., Wang E. S., Huskey N. E., Lim L., Truitt M., McManus M. T., Ruggero D., Goga A., Papa F. R., Oakes S. A. (2012). IRE1alpha cleaves select microRNAs during ER stress to derepress translation of proapoptotic Caspase-2. Science 338, 818–822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ventura A., Young A. G., Winslow M. M., Lintault L., Meissner A., Erkeland S. J., Newman J., Bronson R. T., Crowley D., Stone J. R., et al. (2008). Targeted deletion reveals essential and overlapping functions of the miR-17 through 92 family of miRNA clusters. Cell 132, 875–886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F., Fisher S. A., Zhong J., Wu Y., Yang P. (2015a). Superoxide Dismutase 1 in vivo Ameliorates Maternal Diabetes-Induced Apoptosis and Heart Defects through Restoration of Impaired Wnt Signaling. Circulation. Cardiovasc Genet. 8, 665–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F., Reece E. A., Yang P. (2015b). Advances in revealing the molecular targets downstream of oxidative stress-induced proapoptotic kinase signaling in diabetic embryopathy. Am. J. Obstet. Gynecol. 213, 125–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F., Reece E. A., Yang P. (2015c). Oxidative stress is responsible for maternal diabetes-impaired transforming growth factor beta signaling in the developing mouse heart. Am. J. Obstet. Gynecol. 212, 650 e1–650 e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F., Weng H., Quon M. J., Yu J., Wang J. Y., Hueber A. O., Yang P. (2015d). Dominant negative FADD dissipates the proapoptotic signalosome of the unfolded protein response in diabetic embryopathy. Am. J. Physiol. Endocrinol. Metab. 309, E861–E873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F., Wu Y., Gu H., Reece E. A., Fang S., Gabbay-Benziv R., Aberdeen G., Yang P. (2015e). Ask1 gene deletion blocks maternal diabetes-induced endoplasmic reticulum stress in the developing embryo by disrupting the unfolded protein response signalosome. Diabetes 64, 973–988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F., Wu Y., Quon M. J., Li X., Yang P. (2015f). ASK1 mediates the teratogenicity of diabetes in the developing heart by inducing ER stress and inhibiting critical factors essential for cardiac development. Am. J. Physiol. Endocrinol. Metab. 309, E487–E499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Rong Y. P., Malone M. H., Davis M. C., Zhong F., Distelhorst C. W. (2006). Thioredoxin-interacting protein (txnip) is a glucocorticoid-regulated primary response gene involved in mediating glucocorticoid-induced apoptosis. Oncogene 25, 1903–1913. [DOI] [PubMed] [Google Scholar]
- Watanabe T., Otsu K., Takeda T., Yamaguchi O., Hikoso S., Kashiwase K., Higuchi Y., Taniike M., Nakai A., Matsumura Y., et al. (2005). Apoptosis signal-regulating kinase 1 is involved not only in apoptosis but also in non-apoptotic cardiomyocyte death. Biochem. Biophys. Res. Commun. 333, 562–567. [DOI] [PubMed] [Google Scholar]
- Widmann C., Gibson S., Jarpe M. B., Johnson G. L. (1999). Mitogen-activated protein kinase: conservation of a three-kinase module from yeast to human. Physiol. Rev. 79, 143–180. [DOI] [PubMed] [Google Scholar]
- Wu Y., Wang F., Reece E. A., Yang P. (2015). Curcumin ameliorates high glucose-induced neural tube defects by suppressing cellular stress and apoptosis. Am. J. Obstet. Gynecol. 212, 802 e1–802 e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao L., Cui Y. H., Rao J. N., Zou T., Liu L., Smith A., Turner D. J., Gorospe M., Wang J. Y. (2011). Regulation of cyclin-dependent kinase 4 translation through CUG-binding protein 1 and microRNA-222 by polyamines. Mol. Biol. Cell 22, 3055–3069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang P., Li X., Xu C., Eckert R. L., Reece E. A., Zielke H. R., Wang F. (2013). Maternal hyperglycemia activates an ASK1-FoxO3a-caspase 8 pathway that leads to embryonic neural tube defects. Sci. Signal. 6, ra74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang P., Reece E. A., Wang F., Gabbay-Benziv R. (2015). Decoding the oxidative stress hypothesis in diabetic embryopathy through proapoptotic kinase signaling. Am. J. Obstet. Gynecol. 212, 569–579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ylinen K., Aula P., Stenman U. H., Kesaniemi-Kuokkanen T., Teramo K. (1984). Risk of minor and major fetal malformations in diabetics with high haemoglobin A1c values in early pregnancy. Br. Med. J. 289, 345–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoi T., Fukuo K., Yasuda O., Hotta M., Miyazaki J., Takemura Y., Kawamoto H., Ichijo H., Ogihara T. (2006). Apoptosis signal-regulating kinase 1 mediates cellular senescence induced by high glucose in endothelial cells. Diabetes 55, 1660–1665. [DOI] [PubMed] [Google Scholar]
- Zhang Q., Zhang G., Meng F., Tian H. (2003). Biphasic activation of apoptosis signal-regulating kinase 1-stress-activated protein kinase 1-c-Jun N-terminal protein kinase pathway is selectively mediated by Ca2+-permeable alpha-amino-3-hydroxy-5-methyl-4-isoxazolepropionate receptors involving oxidative stress following brain ischemia in rat hippocampus. Neurosci. Lett. 337, 51–55. [DOI] [PubMed] [Google Scholar]
- Zhong J., Reece E. A., Yang P. (2015). Punicalagin exerts protective effect against high glucose-induced cellular stress and neural tube defects. Biochem. Biophys. Res. Commun. 467, 179–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang R., Rao J. N., Zou T., Liu L., Xiao L., Cao S., Hansraj N. Z., Gorospe M., Wang J. Y. (2013). miR-195 competes with HuR to modulate stim1 mRNA stability and regulate cell migration. Nucleic Acids Res. 41, 7905–7919. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.