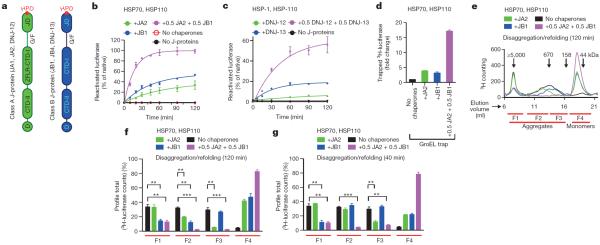
Figure 1. Simultaneous presence of class A and B J-proteins unleashes protein disaggregation activity and broadens target aggregate range of the HSP70 machinery.
a, Two distinct classes (A and B) display highly conserved domain organization involving the HSP70-intertacting HPD motif (red) containing amino-terminal J-domain (JD), Gly/Phe-rich flexible region (G/F), C-terminal β-sandwich domains (CTD-I and II), with class A J-proteins distinguished mainly by a zinc-finger-like region (ZFLR) that inserts into the CTD-I subdomain and a dimerization domain (D)9,23. CTD together with ZFLR provide substrate specificity24,25. b, Disaggregation and reactivation of preformed luciferase aggregates using human HSP70–HSP110 with human J-proteins JA2 (green), JB1 (blue), JA2+JB1 (magenta) or with no J-proteins (black) (n=3). c, Reactivation of heat-aggregated luciferase by nematode HSP70 machinery containing HSP-1, HSP-110 and either alone or in combination with the nematode J-proteins DNJ-12 (A) and DNJ-13 (B) (n=2). d, Fold change in trapped luciferase; control, GroELD87K without other chaperones (black). Values normalized to total 3H counts in each reaction (n=2). e, SEC profile after disaggregation/refolding (120 min) with either J-protein alone or combined. Elution fractions labelled F1–F4 (red lines); F4, disaggregated monomers (~63 kDa). f, Aggregate quantification for fractions F1–F4 from the SEC profile in e. Disappearance of 3H-luciferase from aggregates (F1–F3) occurs with concomitant accumulation of disaggregated monomer (F4). g, Aggregate quantification, after 40-min disaggregation. Values normalized to total counts in each reaction. Two-tailed t-test, **P<0.01, ***P<0.001 (n=3). Data are mean±s.e.m. Precise concentrations are shown in Extended Data Table 1.