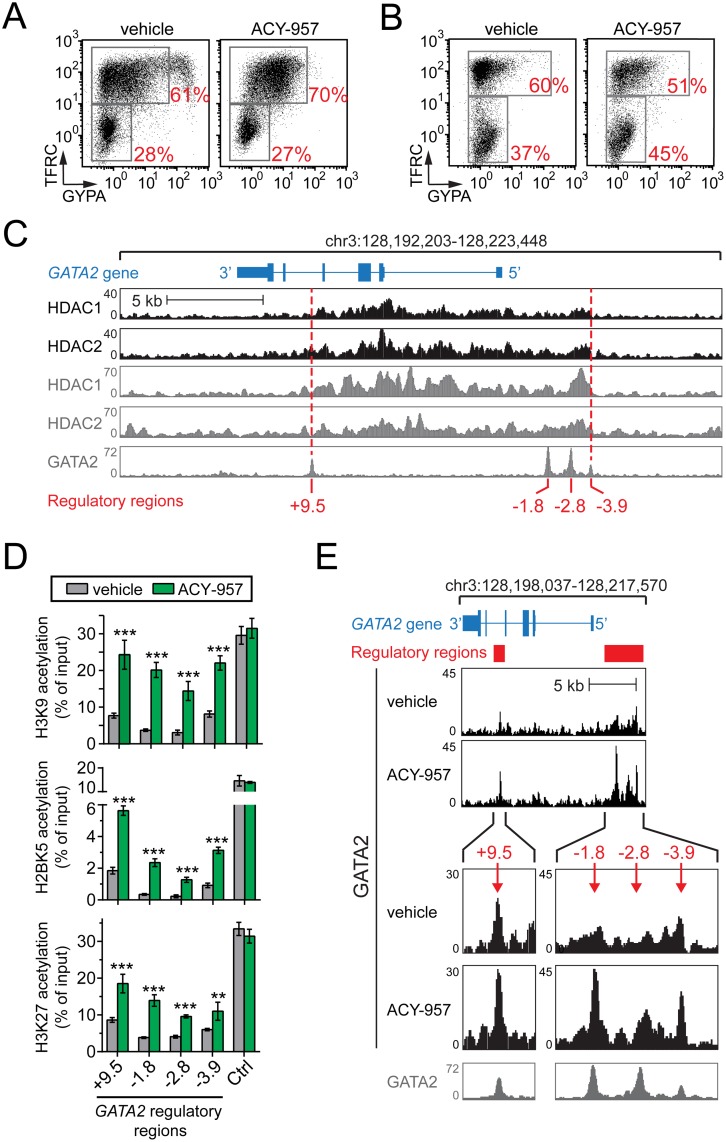
Fig 8. HDAC1/2 inhibition by ACY-957 leads to elevated levels of histone acetylation and GATA2 binding at GATA2 regulatory regions.
BM cells were cultured in CS1 expansion media and then shifted to CS1 differentiation media with vehicle or 1 μM ACY-957 until reaching a differentiation stage similar to cells used in the GeneChip experiments of Fig 5. (A) Erythroid differentiation stage of cells used for ChIP in Fig 8C, 8E and Fig 9A. (B) Erythroid differentiation stage of cells used for ChIP in Fig 8D and Fig 9B. (C) HDAC1 and HDAC2 ChIP-Seq profiles at the GATA2 locus in erythroid progenitors. Chromatin was immunoprecipitated and sequenced using antibodies against HDAC1 or HDAC2 (black histogram tracks). Sequencing read count (y-axis) is plotted as a function of genomic region bin (x-axis). Publically available ENCODE Consortium ChIP-Seq data for HDAC1, HDAC2, and GATA2 in K562 cells is shown (gray histogram tracks) [49, 50]. The previously described GATA2 enhancer regions (red text) map to GATA2 binding peaks in K562 cells. In both K562 and primary erythroid progenitors, HDAC1 and HDAC2 occupy a region bounded by the +9.5 kb and -3.9 kb enhancer regions (red dashed lines). (D) Histone acetylation at GATA2 regulatory regions in vehicle and ACY-957 treated erythroid progenitors. Chromatin from each treatment was immunoprecipitated using anti-H3K9ac, anti-H2BK5ac, or anti-H3K27ac antibodies. A region near the INO80 gene (Ctrl), identified as a region of saturated acetylation across a wide variety of ENCODE cell lines, was used as a control (mean ± SD, n = 2 QPCR replicates for each of n = 3 IP replicates per antibody). P-values were calculated using a two-tailed t test. **P<0.005 and ***P<0.0005 for ACY-957 compared to vehicle control. (E) GATA2 binding at GATA2 regulatory regions following ACY-957 treatment. IP with anti-GATA2 antibody and sequencing as described in ‘C’. ACY-957 treatment resulted in increased GATA2 occupancy (black histogram tracks) at the GATA2 regulatory regions, indicated by solid red bars (wide view) or red arrows (magnified view). ACY-957-responsive regions localize to GATA2 binding peaks in K562 cells (gray histogram track).