FIGURE 2.
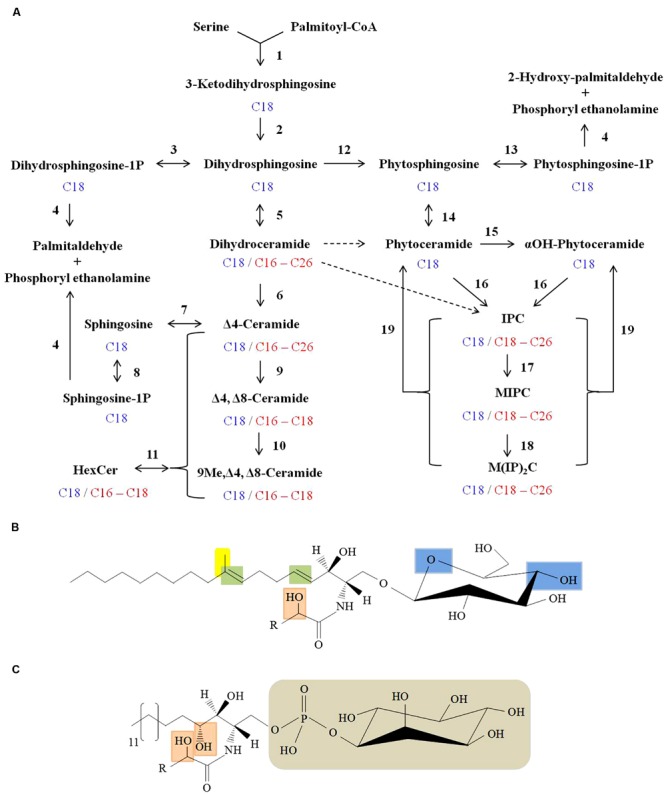
Sphingolipid biosynthesis in fungi. (A) Scheme of fungal sphingolipid biosynthetic pathway. Enzymatic steps with reverse reaction shown in brackets are as follows: (1) Serine C-palmitoyl transferase; (2) 3-ketosphinganine reductase; (3) Dihydrosphingosine kinase [Dihydrosphingosine-1P-phosphatase]; (4) Long-chain base (LCB) phosphate lyase; (5) Dihydroceramide synthase; (6) Dihydroceramide C4 desaturase; (7) Ceramidase [Ceramide synthase]; (8) Sphingosine kinase [Sphingosine-1P-phosphatase]; (9) Ceramide C8 desaturase; (10) Sphingolipid C9 methyltransferase (Smt1); (11) Hexosyl ceramide synthase [Hexosyl ceramidase]; (12) Dihydrosphingosine C4-hydroxylase (13) Phytosphingosine kinase [Phytosphingosine -1P-phosphatase]; (14) Ceramide synthase [Ceramidase]; (15) Sphingolipid α-hydroxylase; (16) IPC synthase; (17) IPC mannosyl transferase; (18) Inositolphosphotransferase; (19) Inositol phosphosphingolipid phospholipase C. Major sphingoid backbone in fungi is composed of 18 C-atoms (shown in blue) and the fatty acyls attached to various sphingolipids comprise of 16–26 C-atoms (shown in red). Putative conversion steps of dihydroceramide to phytoceramide and to IPC are represented by dashed arrows. (B) Structure of fungal glucosylceramide. Long chain sphingoid backbone amide linked to a fatty acyl and linked by β-glycosidic bond to a polar head group (glucose) at C1 position. Unique features of fungal glucosylceramide are: Δ4 and Δ8 double bonds in the sphingoid backbone (shown in green), 9-methylation in the sphingoid backbone (shown in yellow), α-hydroxyl fatty acyl (shown in orange) and hydroxyl groups in the hexose moiety (shown in blue). (C) Structure of fungal IPC. The distinguished features of IPC’s in fungi are: C3-hydroxylation of sphingoid backbone and α-hydroxyl fatty acyl (both shown in orange), phosphate linked inositol group (shown in grey). ‘R’ represents 16–24 carbons in the fatty acyl chain.
