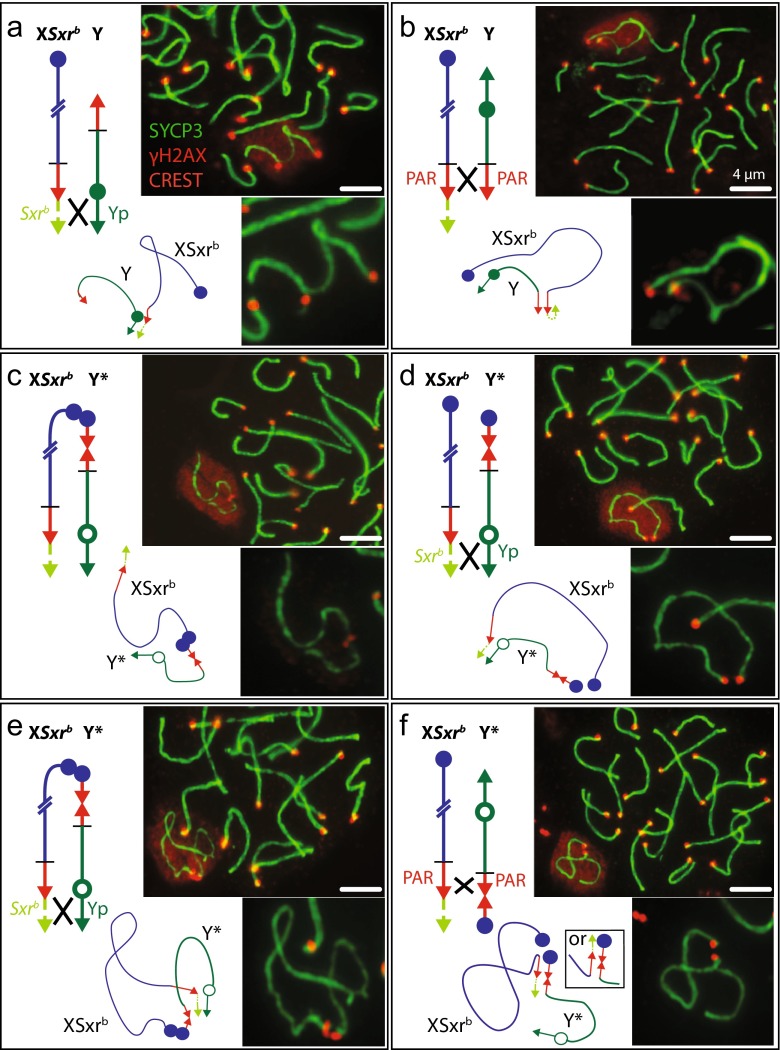
Fig. 4.
X-Y bivalent configurations at pachytene in XSxr bY and XSxr bY* males. Each panel has a diagram of the predicted crossover event, a low magnification view showing the X-Y bivalent in a γH2AX-stained (red) sex chromatin cloud together with near-by autosomes, a higher power view of the X-Y bivalent with the γH2AX staining removed, and a drawing of the deduced X-Y bivalent configuration highlighting the centromere positions. Note that the red CREST staining of active centromeres often appears as yellow where it overlaps the green chromosome axis. (a) XSxr bY with Yp-Sxr b synapsis. (b) XSxr bY with PAR-PAR synapsis. (c) XSxr bY* with centromere-centromere association—this is likely to be due to synapsis driven by homology of Sstx sequences adjacent to the X and Y*(X-derived) centromeres. (d) XSxr bY* with Yp-Sxr b synapsis. (e) XSxr bY* with centromere-centromere and Yp-Sxr b synapsis. (f) XSxr bY* with PAR-PAR synapsis—parasynapsis and staggered synapsis cannot be differentiated at this resolution. The staggered configuration is indicated in the black square