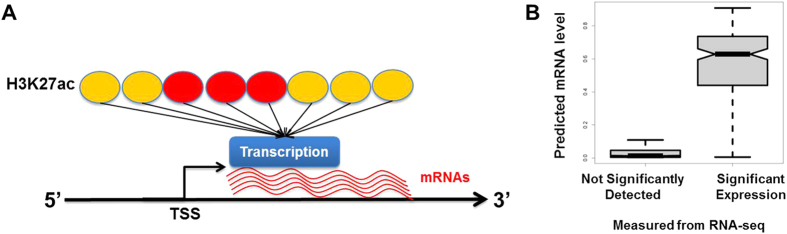
Figure 2. H3K27ac profile around TSS predicts gene regulation in mESC.
(A) A model predicting the mRNA level from H3K27ac ChIP-seq data. H3K27ac ChIP-seq reading signals in 1 kbp around transcription start sites are used to develop the model inferring the mRNA levels from H3K27ac profiles in the TSS regions: 1 and 0 as significant and not significant transcription detected from RNA seq experiments. H3K27ac in the 5′ end of coding regions (red ovals) are more weighted than other H3K27ac (yellow ovals) in the regression model. (B) The predicted mRNA level from the H3K27ac profile around TSS highly correlates with experimental measurements using RNA-seq analysis in mESC. The vertical axis is the predicted mRNA level of the model. The left boxplot shows the distribution of the predicted mRNA levels in genes without significant transcription detected and the right boxplot shows the distribution of predicted mRNA level in genes with significant transcription detected from RNA-seq experiment. The predicted mRNA levels in genes showing significant expression in RNA-seq experiment are significantly higher than those of not significantly detected mRNAs. Wilcoxon rank sum test between significantly expressed and not significantly detected, W = 255195.5, p-value < 10−22.