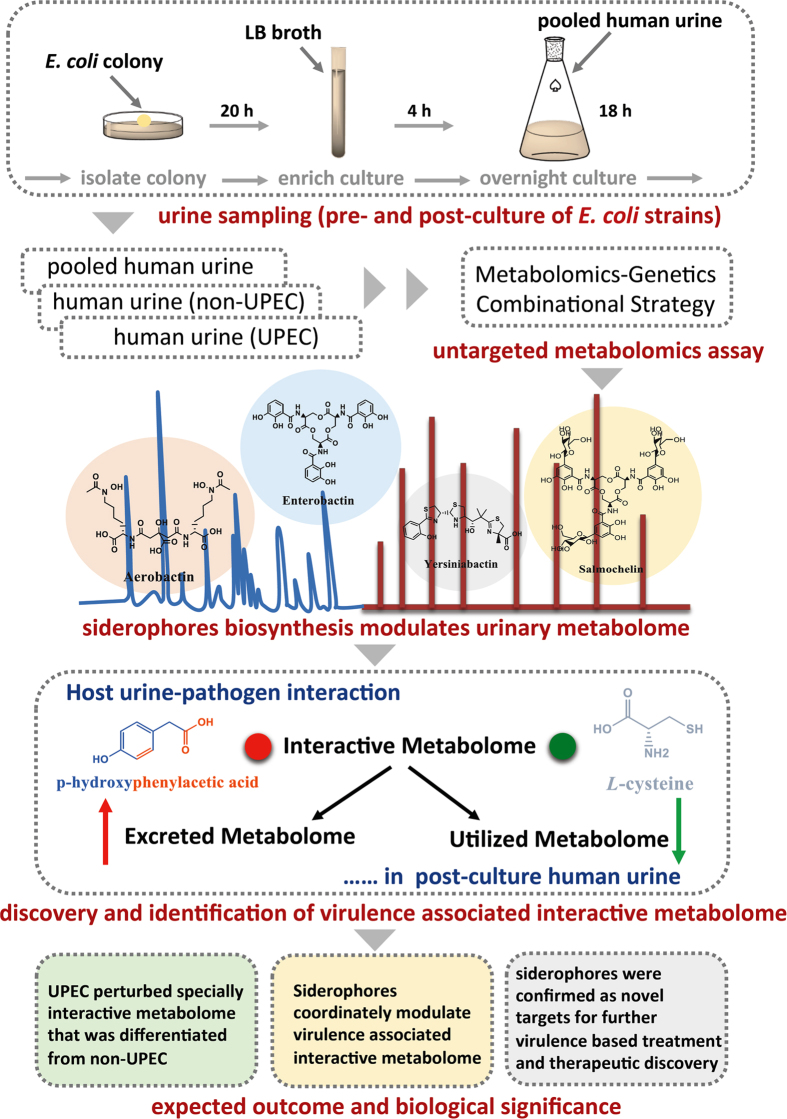
Figure 1. Schematic illustration of the strategy utilized in this study.
Briefly, the sampling procedure was manipulated to incubate all the defined strains with the pooled human urine samples, and then an NMR-based metabolomics analysis was performed to discover the individualized interactive metabolome (utilized metabolome and excreted metabolome) between UPEC and human urine that can differentiate UPEC from non-UPEC. A combination of metabolomics and genetics was employed to elucidate how the siderophores coordinately modulated the virulence-associated interactive metabolome. Finally, our data provide novel insights into the UPEC-human urine interaction and show that siderophore biosynthesis has substantial modulatory effects on the interactive metabolome as the critical components of UPEC virulence. Siderophores may be novel targets for further chemical interventions and the discovery of drugs to treat UPEC-induced urinary tract infection.