Figure 2.
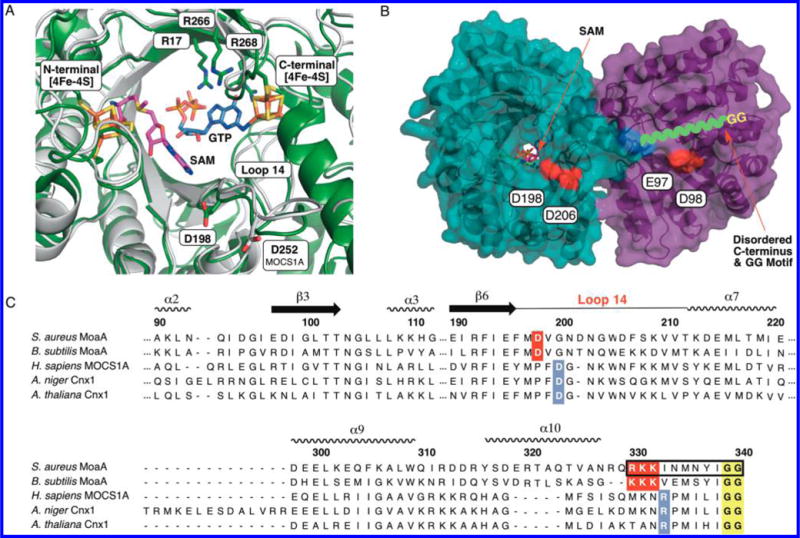
C-terminal GG motif in MoaA and MOCS1A. (A) Overlay of the crystal structure of MoaA in complex with GTP (blue sticks, PDB ID: 2FB3),14 and a homology model structure of MOCS1A (gray). The position of SAM (pink sticks) is based on the crystal structure of MoaA in complex with SAM (PDB ID: 1TV8).10 The homology model of MOCS1A was prepared on the basis of the reported MoaA structure (PDB ID: 1TV8) using SWISS-MODEL.15 Shown in sticks are the active-site residues frequently found mutated in MoCD patients (“hot spots”; Cys ligands of the two 4Fe-4S clusters and three active-site Arg). Also shown are D198, which is essential for the C-terminal tail interaction in S. aureus MoaA, and D252, its putative counterpart in human MOCS1A. (B) S. aureus MoaA homodimer in complex with SAM.10 The disordered C-terminal tail is drawn as a green wavy line, the last ordered residue, Q329, is highlighted in blue, and the acidic residues investigated for the C-terminal tail binding are highlighted in red. (C) Sequence alignment of MoaA/MOCS1A homologues. The GG motif is highlighted in yellow, the C-terminal tail/Loop 14 interface residues in bacteria are highlighted in red, and those predicted for eukaryotes are highlighted in blue. The C-terminal tail sequence corresponding to the 11-mer peptide is indicated in a black square.
