Figure 6.
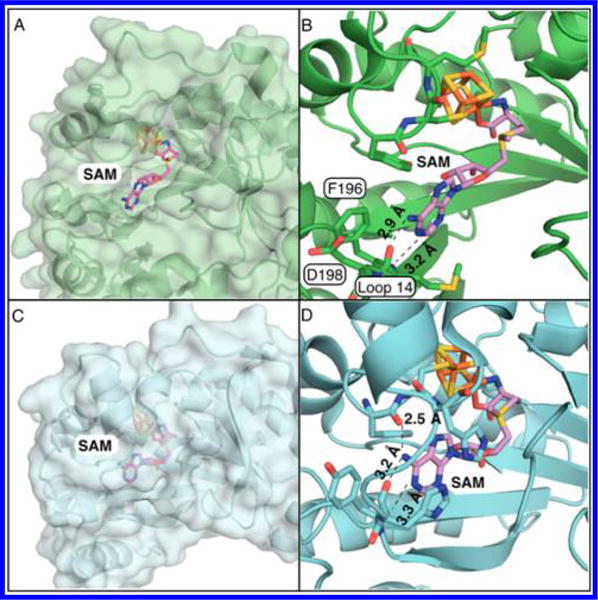
Comparison of active-site structures of MoaA and pyruvate formate-lyase activating enzyme (PFL-AE). (A,B) The active site of S. aureus MoaA in surface representation (A) and in stick and cartoon representation (B).10 (C,D) The active site of PFL-AE in surface representation (C) and in stick and cartoon representation (D).18 (B) and (D) show H-bonding distances (Å) between the adenine of SAM and amino acid residues.
