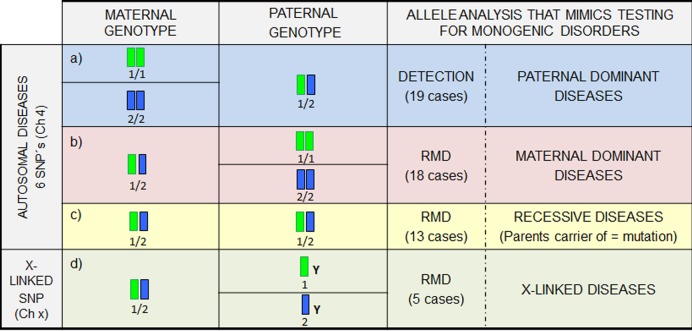
Fig 1. Schematic representation of the study design based on a SNP study showing: parental genotypes, the data analysis approach in maternal plasma and the inheritance pattern mimicked.
Parental genotyping combination for allele 1/allele 2 of a certain SNP can be used to represent an inheritance pattern for a point mutation. The different parental genotype combinations and their correspondence with a specific inheritance pattern are detailed in the figure. Analysis of exclusive paternal sequences requires a detection approach while the study of maternal genomic regions requires a Relative Mutation Dosage (RMD) analysis. A simulation of an NIPD for a dominant disease with paternal origin can be done using a case in which the mother is homozygous for an SNP and the father is heterozygous (blue background). Presence/absence of the exclusive paternal allele could be associated with a fetal genotype. On the contrary, a heterozygous mother and a homozygous father will mimic a dominant disorder with a maternal origin (red background). This simulation can be also considered for recessive diseases in which both parents are carriers of a different mutation. NIPD for a recessive disease in which both parents are carriers of the same mutation can be simulated by taking as an example a couple in which the mother and the father are heterozygous for an SNP (yellow background). Finally, the simulation of NIPD for X-linked disorders can be done by a case in which mother is heterozygous for a SNP and the father is hemizygous (green background).