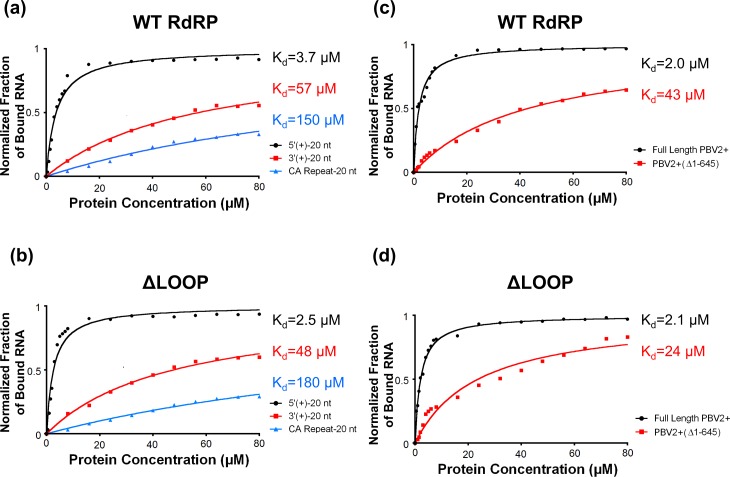
Fig 7.
RNA binding by the hPBV WT (a, c) and ΔLOOP (b, d) RdRPs analyzed by gel shift assays. The fraction of RNA bound was quantitated and plotted as a function of protein concentration. The values were normalized by the total amount of RNA. (a, c) Three distinct RNA oligonucleotides were used, including: (1) the first 20 nucleotides of the 5’-(+)UTR of hPBV genome segment 2 (black); (2) the last 20 nucleotides of the 3’-(+)UTR of the hPBV genome segment 2 (red); and (3) a 20-mer nonsensical CA repeat (blue). (b, d) Two RNA molecules derived from the PBV2+ were used, including the full-length PBV2+ and the PBV2+(Δ1–645). The obtained dissociation constants (Kd) for the different RNA oligonucleotides are indicated on the right.