Summary
HipA is a bacterial serine/threonine protein kinase that phosphorylates targets to effect persistence and multidrug tolerance. HipA is Autophosphorylation of residue Ser150 is a critical regulatory mechanism of HipA function. Intriguingly, Ser150 is not located on the activation loop as in other kinases but in the protein core where it forms part of the ATP-binding “P-loop motif”. How this buried residue is phosphorylated and regulates kinase activity is unclear. Here we report multiple structures revealing that the P-loop motif exhibits a remarkable “in-out” conformational equilibrium, which allows access to Ser150 and its intermolecular autophosphorylation. Phosphorylated Ser150 stabilizes the “out-state”, which inactivates the kinase by disrupting the ATP binding pocket. Thus, our data reveal a heretofore-unseen mechanism of protein kinase regulation that is vital for multidrug tolerance and persistence as kinase inactivation provides the critical first step to allow dormant cells to revert to the growth phenotype and to reinfect the host.
Graphical Abstract
Introduction
Bacterial persistence is largely responsible for the inability of antibiotics to eliminate infections and is caused by a small subpopulation of cells called persisters (Lewis, 2007; Lewis, 2010). Persisters are not mutants but phenotypic variants of wild type cells that escape antibiotic eradication by adopting a transient dormant state (Lewis, 2007; Lewis, 2010). Dormancy provides protection because antibiotics typically act by corrupting active targets into producing toxic by-products. If the cells remain quiescent until the drug is removed and then revert back to growth, multidrug tolerance (MDT) results. A critical component in the development of MDT is the ability of persister cells to emerge from dormancy and re-establish an active growth phase. How such reversion occurs is unknown and represents an outstanding question in the field. The recent discovery of persisters in biofilms has highlighted the importance of understanding these mechanisms as they play important roles in the etiology of recalcitrant infectious diseases. An estimated 65–90% of all infections are associated with biofilms (Lazãr and Chifiriuc, 2010). The phenomenon of MDT, however, has been extremely difficult to study, as persisters comprise less than 1% of a typical bacterial population (Lewis, 2007). In fact, although MDT was first described in 1944 by Bigger, the first bone fide persistence factor was only identified in 1983. This factor was called HipA, for high persistence factor A (Bigger, 1944; Moyed and Betrand, 1983; Moyed and Broderick, 1986).
HipA is a 50 kDa protein, which when mutated led to dramatic increases in persistence in Escherichia coli (Moyed and Bertrand, 1983; Moyed and Broderick, 1986; Black et al., 1991; Black et al., 1994). Subsequently HipA has been shown to be present in multiple Gram-negative bacteria for which genomic sequences are available. In these bacteria, HipA is part of a chromosomally encoded operon, hipBA, which also encodes the 10 kDa HipB protein (Moyed and Bertrand, 1983; Moyed and Broderick, 1986). HipB is a DNA-binding protein that autoregulates transcription of the hipBA operon. HipB also interacts with HipA to disable its persistence function (Black et al., 1991; Black et al., 1994). Recent studies revealed that HipA adopts a eukaryotic serine/threonine protein kinase fold and phosphorylates in vitro the essential translation factor, EF-Tu, suggesting a mechanism of cell stasis via inhibition of protein synthesis (Correia et al., 2006; Evdokimov et al., 2009; Schumacher et al., 2009). The HipA-HipB-DNA structure indicated that HipB neutralizes HipA through conformational inactivation and sequestration (Schumacher et al., 2009). Thus, HipA and HipB can be categorized as a chromosomally encoded toxin-antitoxin (TA) module, where HipA is the toxin and HipB is the antitoxin (Hayes and Van Melderen, 2011). However, neither HipA nor HipB shows homology with any of the TA proteins in any system marking them as a unique TA module.
In addition to its binding to HipB, HipA is regulated by autophosphorylation on residue Ser150 (Correia et al., 2006). Remarkably, the HipA structures reveals that Ser150 is buried in the protein core (Schumacher et al., 2009). This is in sharp contrast to the phosphorylated regulatory sites of other protein kinases, which are typically located on solvent exposed loops. Studies in which Ser150 was replaced by alanine resulted in a loss of persistence, which led to the suggestion that Ser150 phosphorylation is necessary for kinase activation and therefore, persistence (Correia et al., 2006). To gain insight into the seemingly paradoxical questions of how a residue buried in the protein hydrophobic core can be phosphorylated and how this phosphorylation affects HipA kinase activity, we carried out structural and biochemical studies on this key persistence factor. The data reveal a heretofore-unseen mechanism of kinase regulation and suggest a molecular basis for how persister cells can emerge from dormancy to cause reinfection.
Results and Discussion
Structure determination of pHipA and comparison with nonphosphorylated HipA
To gain insight into how the buried Ser150 residue in HipA is phosphorylated we purified the wild type (wt) phosphorylated HipA protein (pHipA) and performed structural studies. Two crystal structures (trigonal and monoclinic) of pHipA were determined to 1.90 Å and 1.50 Å resolution, respectively, by molecular replacement using the previously determined structure of HipA(D309Q), a catalytically inactive enzyme due to the replacement of the catalytic base, residue Asp309, with a glutamine (Jones et al., 1991; Brünger et al., 1998; Schumacher et al., 2009). The final Rwork/Rfree values for the structures were 18.4%/24.3% and 21.2%/24.8%, respectively (Table S1).
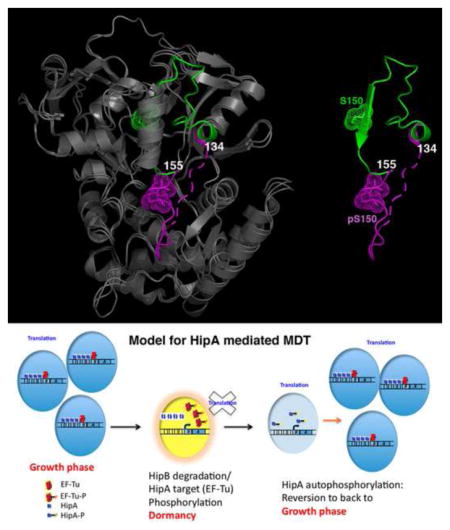
Comparison of the pHipA and nonphosphorylated HipA(D309Q) structures showed that whilst they are similar overall, the region of pHipA, which encompasses residues 134–155 and includes phosphorylated Ser150 (pS150), adopts a vastly different conformation (Figure 1A–C). In the HipA(D309Q) structure residues 134–155 are part of the hydrophobic core of the N-domain. In the pHipA structures these residues are extruded by over 30 Å from their internal location into the solvent. In the monoclinic pHipA structure, residues 143–155 are mostly disordered whilst in the trigonal pHipA structure these residues form a well-defined extended structure that lies over the active site and interacts with the putative activation loop/region (residues 185–195), which is disordered in the HipA(D309Q) structure (Schumacher et al., 2009). Two flexible regions consisting of residues 134–147 and 155–157 appear to provide the necessary conformational malleability to permit loop extrusion (Figure 1A). Also important for loop ejection is the paucity of contacts between HipA residues 148–154 and other residues in the protein core and a repositioning of helix α3 (Supplementary Movie 1).
Figure 1. Extrusion of the P-loop motif of Ser150-phosphorylated HipA (pHipA).
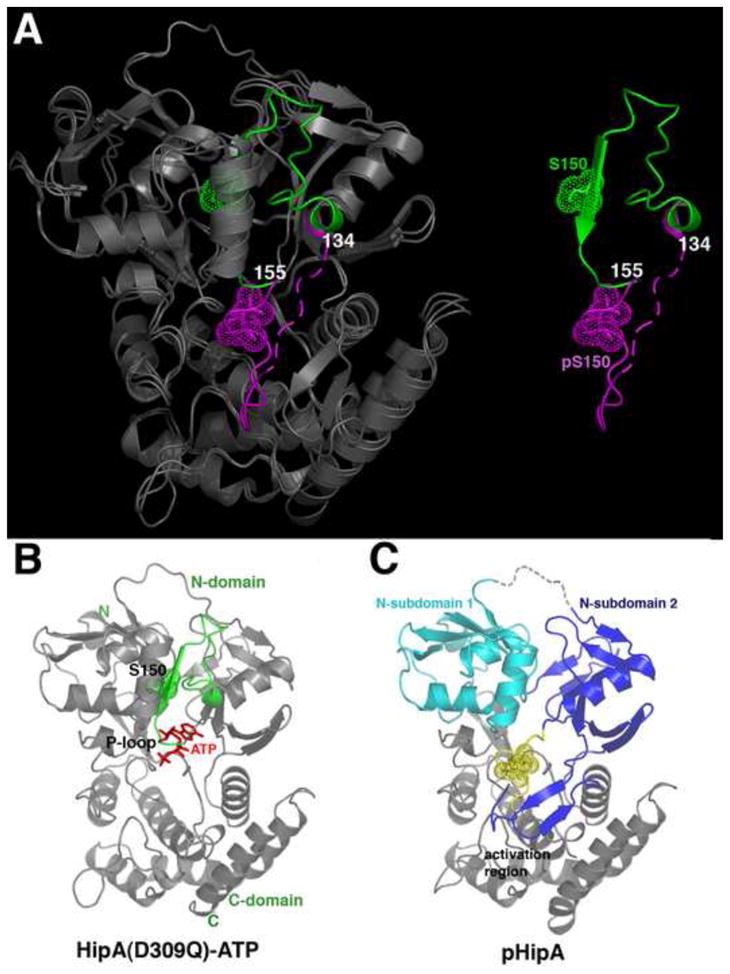
(A) Superimposition of HipA(D309Q) and pHipA. (Left) The structures are shown as ribbons and colored grey except for their P-loop motifs, which are green in HipA(D309Q) and magenta in pHipA. (Right) Only the P-loop motifs are shown to underscore the dramatic conformational change resulting from the ejection of the P-loop motif from the protein core. Phosphorylated Ser150 (pS150) and Ser150 (S150) are shown as dotted spheres. Residues 134 and 155 are also labelled for reference.
(B) Ribbon diagram of the HipA(D309Q)-ATP structure (PDB code, 3DNU) (Schumacher et al, 2009). The P-loop motif is colored green and ATP is shown as red sticks.
(C) Ribbon diagram of pHipA in the same orientation as Figure 1B highlighting the separate N-subdomains (colored cyan and blue) and folded activation region that are created by P-loop motif ejection. pS150 is shown as dotted yellow spheres.
Destruction of the HipA catalytic P-loop motif by Ser150 expulsion: the in and out states
The finding that a portion of the protein core is ejected and becomes solvent exposed in pHipA is striking, but more critical is the finding that these expelled residues encompass a catalytically essential region, the so-called catalytic glycine rich or P-loop motif (Fernandez-Fuentes et al., 2004; Taylor and Kornev, 2011). Typical protein kinase glycine rich motifs consist of an 8 residue hairpin-like loop that is located between two β strands. The tip of this β-loop-β motif (herein referred to as the P-loop motif) encircles the ATP phosphates and is essential for catalysis. In HipA, the P-loop motif (151-VAGAQEKT-158) diverges from those found in other protein kinases as it contains only one glycine (Pereira et al., 2011). As a result it has a slightly altered conformation when compared with typical serine/threonine kinase P-loops. Nonetheless, in the HipA(D309Q)-ATP structure this region is used to embrace the ATP phosphate groups (Figure 1B). What is particularly notable is that the structural change observed in the pHipA structures leads to the complete destruction of this essential catalytic loop conformation (Figure 1A–B).
Thus, the structural data on HipA indicate that the P-loop motif exists in equilibrium between two conformations, which we call the “in-state” and “out-state”. We note that the out-state is not meant to indicate one specific conformation but rather any conformation in which the P-loop motif is ejected from the hydrophobic core. Interestingly, the HipA N-terminal domain in which the P-loop motif resides is larger than the corresponding domains of most protein kinases. In HipA, the P-loop motif forms the central strand of the β-sheet of the domain. Hence, ejection of this region disrupts this β-sheet of the N-domain and leads to the creation of N-subdomains 1 and 2 (Figure 1C). N-subdomain 2 contains substrate-binding residues whilst the role of N-subdomain 1 is unknown but mutations in residues in this subdomain can result in a high persistence phenotype (Moyed and Bertrand, 1983; Korch et al., 2003; Korch and Hill, 2006; Lewis, 2007). The cavity between the N-subdomains that results from P-loop motif expulsion is filled with solvent, which now links the disjointed strands of the N-domain β sheet.
Additional comparison of in-state and out-state structures reveals different sets of contacts in each state and these interactions, which are nearly equal in number, appear to be equally balanced. In the in-state, Arg78 forms multiple hydrogen bonds with the backbone carbonyl oxygens of residues Met140, Ile141 and Glu144. These interactions buttress residues 140 through 147, allowing them to form a distorted helix-loop structure (Figure 2A–B). In the outstate, Tyr132 and Arg78 rotate into the cavity vacated by residues 140–147. From its new location the side chain of N-subdomain 2 residue Tyr132 makes a weak hydrogen bond to N-subdomain 1 residue Tyr79 and residue Arg78 now interacts with the side chain of Glu156, which is found on the tip of the P-loop motif of ATP-bound (in-state) HipA (Figure 2A–B). In order to form this latter contact Glu156 adopts a conformation that disfavors canonical P-loop formation. Thus, these contacts and the nonstandard P-loop conformation aid in the unravelling of the P-loop motif. By contrast, in the ATP-bound state, Glu156 interacts with Arg74 and this interaction favors the ATP-binding conformation of the P-loop motif. Consistent with playing a key role in HipA function, Arg78 is conserved in all HipA proteins.
Figure 2. The phosphorylated P-loop motif can interact with and occlude the HipA catalytic pocket.
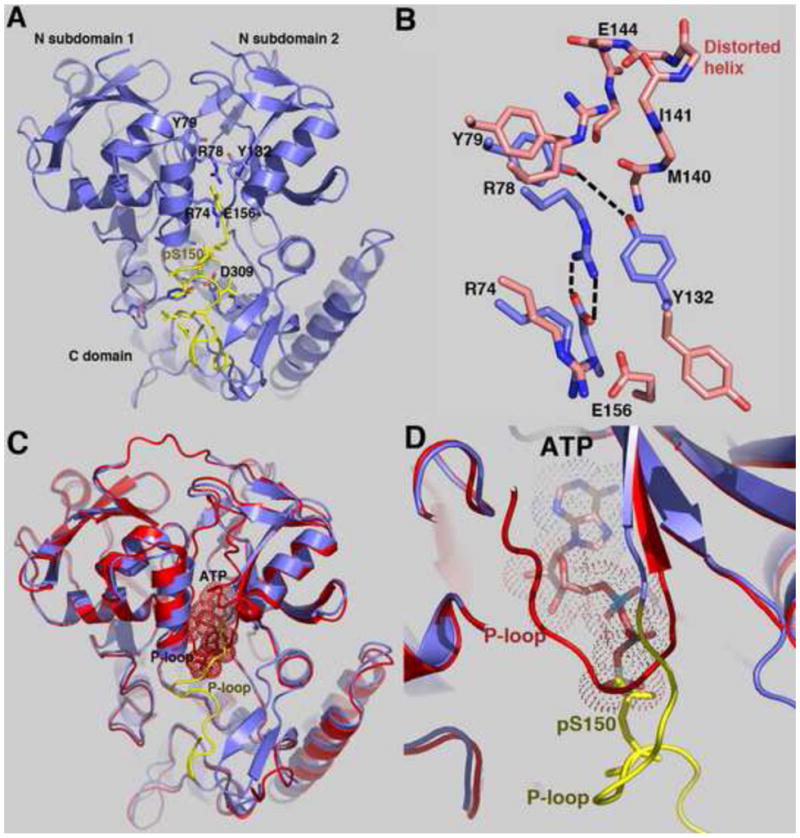
(A) Ribbon diagram of the trigonal pHipA structure. The P-loop motif is colored yellow and residues involved in mediating interactions between N-subdomains 1 and 2 and between the P-loop motif and the protein are shown as sticks and labelled. The catalytic base, residue Asp309 (D309) is labelled.
(B) View of the interactions nearby and involving residue R78 in the out-state (pHipA, blue) and the in-state (HipA(D309Q), salmon) conformations. Also shown are contacts between Y79 and Y132 that help stabilize the “out-state” conformation.
(C) Overlay of the trigonal pHipA (blue) and HipA(D309Q)-ATP (red) structures. The superimposition shows that the phosphate group of pS150 overlaps the ATP γ-phosphate (dotted spheres) in this autoinhibited out-state conformation. Each P-loop is labelled and that belonging to pHipA is coloured yellow.
(D) Close-up of the overlapping ATP γ-phosphate and the pS150 phosphate group. The ATP and phosphate group of pS150 are shown as sticks and dotted surfaces and the P-loop motifs of pHipA (yellow) and HipA(D309Q)-ATP (red) are labelled.
The phosphorylated HipA pS150 loop can function as an intramolecular “product” autoinhibitor
In the trigonal pHipA structure, the P-loop motif binds in the active site and the phosphate of residue pSer150 overlaps the position of the γ-phosphate of ATP observed in the HipA(D309Q)-ATP structure (Figure 2C–D). Moreover, when bound in this manner, the pSer150 side chain interacts directly with the side chain of the proposed catalytic base, residue Asp309. The utilization of a loop, which mimics a bound substrate, is a well-described mechanism to inhibit protein kinases. These so-called “pseudo-substrates” can be provided from regulatory subunits or loops within the kinase itself (Taylor and Radzio-Andzelm, 1997; Cheetham, 2004). In the pHipA structure the phosphorylated Ser150 functions as a novel kind of intramolecular “pseudo-product’ inhibitor that not only comes from the protein itself but is part of the active site. Interestingly, when the P-loop motif binds to the catalytic pocket, residues 134–155 of the P-loop motif interact with and permit folding of the normally disordered activation region (residues 185–195) into a β hairpin (Figure 1C). Folding of the activation region of HipA has been observed previously only when an EF-Tu peptide substrate was bound (Schumacher et al., 2009).
Structural basis of HipA inactivation by the S150A mutation
The ability of the P-loop motif of HipA to take an out-state conformation provides an explanation for autophosphorylation of residue Ser150 as expulsion of the P-loop motif exposes this residue to the solvent (Figure 1A). Once phosphorylated, the large electronegative phosphate group precludes reinsertion of the P-loop motif into the core. However, previous data showing that mutation of residue Ser150 to alanine (S150A) abolishes the ability of HipA to mediate persistence led to the suggestion that phosphorylation of Ser150 is required for kinase activation (Correia et al., 2006). This appears contrary to our structural data, which would predict that the phosphorylated out-state conformation of the HipA would be inactive. To gain insight into the conformational and functional effects of the S150A mutation, we solved the crystal structure of HipA(S150A) to 1.88 Å resolution (Table S1, Figure S1A–B).
Unexpectedly, the HipA(S150A) structure revealed that residues 133–155 are no longer present in the protein hydrophobic core and, similar to the monoclinic pHipA structure, the P-loop motif of HipA(S150A) is largely disordered and points into the solvent. Thus, the P-loop motif of HipA(S150A) adopts the out-state conformation (Figure S1A–B). The underlying reason why the S150A mutation favors the out-state is suggested by the HipA(D309Q) structure, which shows that in the in-state the Ser150 side chain makes a hydrogen bond to the carbonyl oxygen of residue Leu64 (Figure S1C). Because there are few interactions between the loop encompassing Ser150 and the hydrophobic core, the Ser150-Leu64 hydrogen bond is likely a key contributor to the stability of the in-state conformation (Figure 1B). As a result, the loss of this interaction in the S150A protein would favor the out-state conformation or at the least disfavor the in-state conformation.
HipA autophosphorylation prevents ATP binding
Our structural data show that the P-loop motifs of pHipA and HipA(S150A) take the out-state conformation, which we predict would lead to the loss of physiologically relevant ATP binding and hence to an inactive kinase. To test this hypothesis we analysed the ability of pHipA and HipA(S150A) to bind adenine nucleotides. Isothermal titration calorimetry (ITC), employed previously to analyze ATP binding to HipA(D309Q) (Schumacher et al., 2009), revealed no interaction between ATP or the nonhydrolyzable ATP analogue, AMPPNP (adenylylimidodiphosphate), and pHipA. To ensure that this apparent lack of binding was not an experimental artefact we developed an ultraviolet-visible spectroscopy-based thermal denaturation assay, hypothesizing that HipA stability would increase as a function of nucleotide concentration (Waldron et al., 2003; Guedin et al., 2010). (Figure 3A–D). These studies demonstrated a saturable increase in the thermal stability of HipA(D309Q) upon the addition of ATP or AMPPNP consistent with high affinity adenine nucleotide binding (Kd ≤ 100 μM) (Figure 3A) and with our previous ITC studies (Schumacher et al., 2009). By contrast, pHipA showed no increase in its thermal stability even in the presence of 6.0 mM ATP (Figures 3B and S2A). Congruent with its preference for the out-state, HipA(S150A) showed a small shift in its thermal stability but only after the addition of more than 2.0 mM ATP (Figure 3C). As expected, ADP also significantly stabilized dephosphorylated HipA. By contrast, greater than 2.0 mM ADP was required to increase the stability of pHipA (Figure S2B).
Figure 3. Phosphorylation of residue Ser150 inhibits ATP binding and the catalytic activity of HipA.
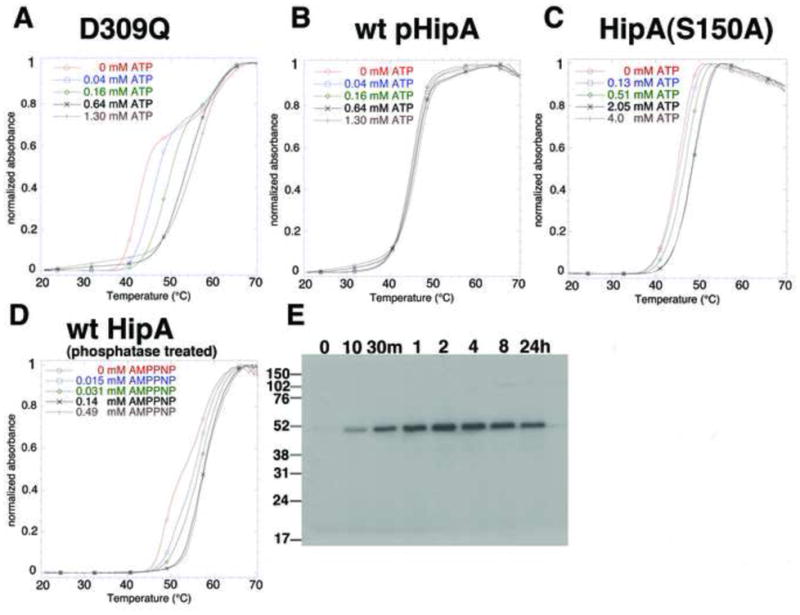
(A) Thermal denaturation of HipA(D309Q) as a function of increasing ATP concentration. The normalized absorbance is plotted as a function of temperature. The concentration range of ATP is given within this panel and panels B–D.
(B) Thermal denaturation of pHipA as a function of increasing ATP concentration.
(C) Thermal denaturation of HipA(S150A) as a function of increasing ATP concentration.
(D) Thermal stability of HipA, which has been dephosphorylated by λ phosphatase, as a function of increasing AMPPNP concentration. Note the low micromolar concentration of AMPPNP (between 31 and 140 μM) needed to complete the thermal shift.
(E) Time course of autophosphorylation of λ phosphatase-treated wild type HipA.
Phosphorylation provides a rapid mechanism for cells to control the activity of a protein and is typically reversible (Taylor and Kornev, 2011). To determine whether or not dephosphorylation of pHipA restores its ability to bind ATP and its kinase activity, pHipA was dephosphorylated by λ phosphatase (Barik et al., 1993). Thermal stability assays demonstrated that this protein was able to bind AMPPNP with high affinity (Kd ~30 μM) (Figure 3D). Thus, these data demonstrate that pHipA and HipA(S150A) do not bind ATP or ADP at physiologically relevant concentrations and support the idea that the HipA(S150A) mutant is inactive because it prefers strongly the out-state conformation.
pHipA cannot bind adenine nucleotides productively
Autophosphorylation of an ejected residue that once formed part of the protein core and active site is unprecedented but whether phosphorylation is intramolecular or intermolecular is unclear. Indeed, the finding from the pHipA structure that the phosphorylated Ser150 residue can dock into the active site with its phosphate moiety overlapping the site of the γ phosphate of bound ATP suggested the possibility that autophosphorylation may be intramolecular. However, an intramolecular phosphorylation reaction would require ATP binding and γ phosphate transfer to Ser150 concomitant with loop expulsion; a stereochemically and kinetically unlikely occurrence. Thus, to examine the affect of P-motif ejection on nucleotide binding, we crystallized pHipA in the presence of 3 mM ADP, as the ligand-induced thermal stability data show that pHipA can bind ADP at elevated concentrations (Figure S2B). The pHipA-ADP structure was determined to 2.30 Å resolution with Rwork/Rfree values of 23.8%/28.0%. As a control, the HipA(D309Q)-ADP structure was solved to 2.30 Å resolution and refined to Rwork/Rfree values of 22.1%/26.3% (Table S1).
Strong electron density was observed for the P-loop motif and the entire ADP molecule in the HipA(D309Q)-ADP structure (Figure 4A). In addition to van der Waals contacts to the ribose moiety, the P-loop motif of the HipA(D309Q) embraces the ADP nucleotide phosphates whereby the amide nitrogens of P-loop residues Ala154 and Gln155 interact with the ADP β phosphate whilst the Lys157 side chain contacts both the α and β phosphates (Figure S1C). By sharp contrast, the structure of the pHipA-ADP complex shows the P-loop motif takes the outstate, as observed in other pHipA structures, despite the presence of nucleotide. More telling, electron density is observed only for the adenine moiety of the ADP (Figure 4A). Thus expulsion of P-loop motif residues from the pHipA catalytic site eliminates key contacts to the ADP ribose and phosphate groups, whilst retaining interaction with the adenine moiety (Figure 4A–B). As noted above, the transfer of the γ phosphate of ATP to the substrate by serine/threonine protein kinases demands not only tight binding of ATP but also the precise alignment of the γ phosphate and substrate hydroxyl group. The lack of density for the ribose and phosphate moieties of ADP in the pHipA-ADP structure provides visible evidence to support the idea that the kinase P-loop motif is required to anchor the phosphate moieties in place for efficient catalysis. Without these stabilizing interactions the phosphates are too flexible and indicate intramolecular autophosphorylation would be highly improbable. Thus, the structural data show that pHipA is unable to bind adenine nucleotides productively and suggests that HipA Ser150 autophosphorylation is an intermolecular event.
Figure 4. HipA phosphorylation is intermolecular: a model for MDT development.
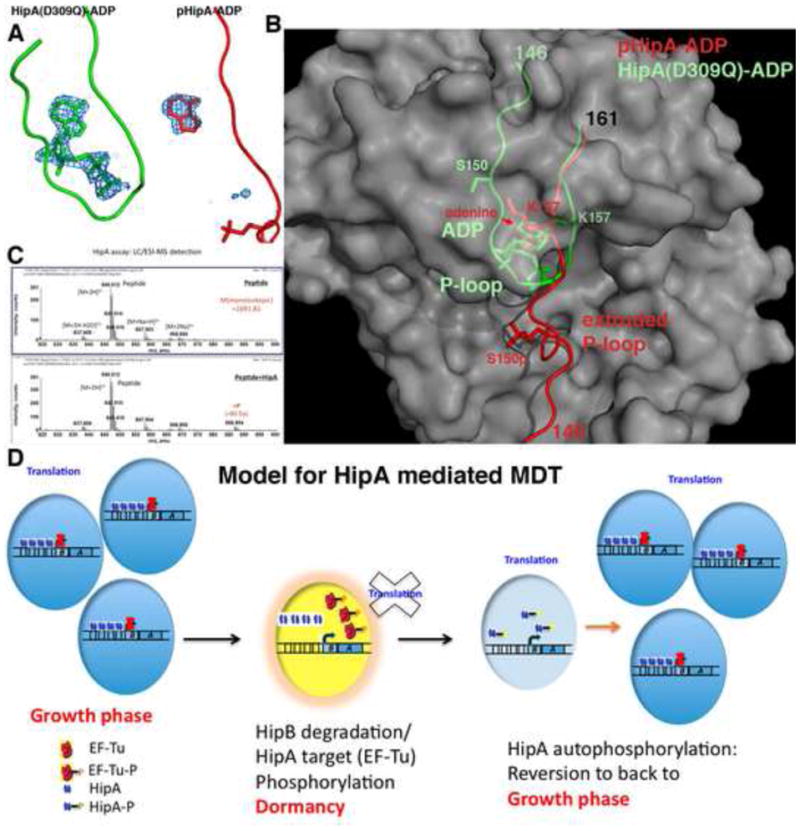
(A) Fo-Fc electron density maps (blue mesh, contoured at 4σ) of the HipA(D309Q)-ADP (left) and pHipA-ADP (right) structures calculated before inclusion of nucleotide. Weak electron density is observed only for the adenine moiety in the pHipA-ADP complex whereas density for the full ADP molecule is seen in the HipA(D309Q)-ADP complex.
(B) View into the catalytic pocket of HipA after superimposition of the HipA(D309Q)-ADP and pHipA-ADP structures. The P-loop motif of the HipA(D309Q)-ADP structure is shown in green and that of the pHipA-ADP structure is shown in red. In the pHipA-ADP structure the P-loop is ejected thereby destroying nearly the entire nucleotide binding pocket whilst in the HipA(D309Q)-ADP structure the P-loop motif is folded into the protein core and forms the canonical P-loop-turn structure that embraces the ADP phosphates. The overlaid HipA proteins are shown as grey surface representations.
(C) Mass spectrometry analysis of an in vitro phosphorylation assay examining the ability of dephosphorylated HipA to phosphorylate the HipA peptide, EENDFRISVAGAQEK, which encompasses HipA residue Ser150. Top frame is the control (no HipA added to the peptide) and the bottom frame included dephosphorylated wtHipA. The presence of the phosphorylated peptide product (+ 80 Da) was clearly evident in the presence of dephosphosphorylated HipA and is denoted by +P in red.
(D) Model for HipA-mediated MDT. HipA normally is inhibited via its interaction with HipB, which binds four operator sites in the hipBA promoter. If enough of the intrinsically labile HipB protein is proteolysed to allow HipA release, HipA will phosphorylate EF-Tu and potentially other targets causing cell stasis (indicated by the yellow cell) (Schumacher et al., 2009). To emerge from dormancy, HipA must be inactivated. This is mediated by the unusual intermolecular autophosphorylation of its P-loop motif. This provides cells time to “reset” and re-enter the growth phase. If the persister cells achieved dormancy during antibiotic treatment and remained quiescent until drug removal, the emergence of cells from the dormant stage leads to reinfection or multidrug tolerance.
HipA autophosphorylation is intermolecular
To address unambiguously whether the phosphorylation of HipA is an intra or intermolecular event we examined the ability of λ phosphatase-dephosphorylated HipA to phosphorylate a peptide corresponding to HipA residues 143–157 (EENDFRISVAGAQEK), which includes Ser150. Mass spectrometry was carried out and revealed that the peptide was indeed phosphorylated by HipA (Figure 4C). This demonstrates that autophosphorylation can take place intermolecularly. However, to test this in vivo we co-expressed HipA and kinase-dead, hexahistidine-tagged-HipA(D309Q) and performed mass spectrometry on the purified D309Q protein. Because HipA(D309Q) shows no kinase activity, phosphorylation of its Ser150 residue would provide clear evidence for in vivo intermolecular phosphorylation by the co-expressed wt HipA protein. This analysis revealed that HipA(D309Q) was, in fact, phosphorylated. Thus, both in vitro and in vivo studies support that HipA autophosphorylation is intermolecular. This finding is consistent with the fact that the P-loop motif of pHipA can no longer bind ATP due to its restructuring and ejection from the protein core and that HipA phosphorylation must be carried out by a second HipA with an instate conformation P-loop. Thus, our combined data show that the pHipA is inactive. Having deduced that HipA phosphorylation is intermolecular, we utilized our kinase assay to assess phosphorylation of the kinase dead HipA(D309Q) by the HipA(S150A) and pHipA proteins. Strikingly, HipA(S150A) showed no kinase activity even when using ATP concentrations as high as 3 mM (Figure S3A). By contrast, the pHipA sample showed a small amount of autophosphorylation. However, this phosphorylation is the direct result of the incomplete modification of the HipA protein. Indeed, mass spectrometry analysis of pure pHipA reveals ~25% of the protein is not phosphorylated (Figure S3B). In accord, increasing the percentage of phosphorylated HipA diminishes the observed but artefactual kinase activity of pHipA.
A novel mechanism for kinase inactivation plays a central role in multidrug tolerance
The data show that HipA utilizes a newly described mechanism of kinase autoinhibition involving P-loop motif ejection followed by intermolecular autophosphorylation. By contrast regulatory phosphorylation of essentially all protein kinases involves modification of activation region/loop residues. CDK2, which is phosphorylated by a Wee1/Mik1 kinase on P-loop residue Tyr15, is the only other example of P-loop residue phosphorylation (Welburn et al., 2007). However, because Tyr15 is located on the exposed P-loop tip and CDK2 possesses a much smaller N-domain than HipA, Tyr15 phosphorylation requires no conformational change and the resultant CDK2-pTyr15 structure is essentially identical to the non-phosphorylated protein (Figure S4A–B). Moreover, CDK2-pTyr15 can still bind ATP and phosphorylation appears to inhibit function simply by blocking the protein substrate-binding site (Welburn et al., 2007). By sharp contrast, residue Ser150 of HipA is buried in the protein core and a dramatic structural change is required to effect its phosphorylation.
Our combined data suggest a HipA-mediated persistence cycle in which HipA is initially inhibited by binding to HipB thereby sequestering HipA from substrates and locking this enzyme in an inactive conformation (Schumacher et al., 2009). Indeed, co-expression of HipB and HipA reveals no HipA-autophosphorylation (Evdokimov et al., 2009). Upon release from HipB, HipA phosphorylates target proteins thereby inducing persistence. Subsequent autophosphorylation is the crucial initial event that is necessary to allow persister cells to revert to a growth phenotype as this modification squelches the HipA kinase activity (Figure 4D). Whether pHipA is degraded ultimately is currently unknown and a difficult issue to address given that only 1 in a million cells become a persister and each persister cell contains only a few molecules of HipA. This dearth also makes tracking how the conformations of a few molecules of HipA contribute to the frequency and duration of persistence currently intractable. Logically pHipA-degradation and HipB binding to newly synthesized HipA would be the most effective subsequent steps to restore the growth phenotype. On the other hand, control of the length of persistence could involve a robust mechanism of dephosphorylation of pHipA. However, our efforts to identify a HipA specific phosphatase have been unsuccessful thus far. Regardless the conformational dynamics of the P-loop motif of HipA dramatically and directly affect persistence as any event, which favours the out-state, including phosphorylation of Ser150 or loss of an in-state stabilizing interaction as observed in HipA(S150A), results in the loss of persistence (Correia et al., 2006).
Finally, a simple bioinformatic analysis of the currently available bacterial genomes reveals that the hipBA system is highly conserved amongst Gram-negative bacteria with HipA proteins showing sequence identities ranging from 45% to 95%. Strikingly, Ser150 is as highly conserved as the catalytic residues of these putative HipA proteins. These findings suggest that the hipBA system is an important MDT/persister module in many Gram-negative organisms and further, that HipA autophosphorylation, which provides a self-inactivation mechanism, plays a fundamental role in MDT development.
Experimental Procedures
HipA proteins were purified via Ni2+-NTA chromatography. Purified wt HipA is typically 65–90% phosphorylated on residue Ser150 (Correia et al., 2006 and our results). Dephosphorylated wt HipA was prepared by treating pHipA with λ phosphatase. pHipA, HipA(S150A), pHipA-ADP and HipA(D309Q)-ADP structures were solved by molecular replacement. Phosphorylation of the HipA 15mer peptide by dephosphorylated HipA or pHipA was analyzed by mass spectrometry. For a full description of all the experimental methods see Extended Experimental Procedures.
Supplementary Material
Highlights.
Dormant bacterial cells called persisters are the origin of multidrug tolerance (MDT)
HipA is an MDT kinase that is regulated by autophosphorylation of residue S150
S150 is the phosphorylated target but is buried in the hydrophobic core
When ejected, S150 is phosphorylated and destroys the catalytic P-loop structure
Acknowledgments
We thank the Advanced Light Source and their support staff. The ALS is supported by the Director, Office of Science, Office of Basic Energy Sciences, Materials Sciences Division, of the US Department of Energy at the Lawrence Berkeley National Laboratory. This work was supported by a U.T. M.D. Anderson Trust Fellowship and National Institutes of Health grant GM074815 (to M.A.S.) and R.A. Welch grant G0040 (to R.G.B.). KL is supported by National Institutes of Health grant 3R01 GM061162-05A1.
Footnotes
Coordinates and structure factor files for HipA(S150A), pHipA P21, pHipA P3121, HipA(D309Q-ADP) and pHipA-ADP have been deposited with the Protein Data Bank under the accession code 3TPB, 3TPD, 3TPE, 3TPT and 3TPV, respectively.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Barik S. Expression and biochemical properties of a protein serine/threonine phosphatase encoded by bacteriophage lambda. Proc Natl Acad Sci USA. 1993;90:10633–10637. doi: 10.1073/pnas.90.22.10633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bigger J. Treatment of staphylococcus infections with penicillin. Lancet. 1944;11:497–500. [Google Scholar]
- Black DS, Irwin B, Moyed HS. Autoregulation of hip, an operon that affects lethality due to inhibition of peptidoglycan or DNA synthesis. J Bacteriol. 1994;176:4081–4091. doi: 10.1128/jb.176.13.4081-4091.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black DS, Kelly AJ, Mardis MJ, Moyed HS. Structure and organization of hip, an operon that affects lethality due to inhibition of peptidoglycan or DNA synthesis. J Bacteriol. 1991;173:5732–5739. doi: 10.1128/jb.173.18.5732-5739.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brünger AT, Adams PD, Clore GM, DeLano WL, Gros P, et al. Crystallography and NMR System: A new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- Cheetham GMT. Novel protein kinases and molecular mechanisms of autoinhibition. Curr Opin Struct Biol. 2004;14:700–705. doi: 10.1016/j.sbi.2004.10.011. [DOI] [PubMed] [Google Scholar]
- Correia FF, D’Onofrio A, Rejtar T, Li L, Karger BL, Makarova K, Koonin EV, Lewis K. Kinase activity of overexpressed HipA is required for growth arrest and multidrug tolerance in Escherichia coli. J Bacteriol. 2006;188:8360–8367. doi: 10.1128/JB.01237-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evdokimov A, Voznesensky I, Fennell K, Anderson M, Smith JF, Fisher DA. New kinase regulation mechanism found in HipAB: a bacterial persistence switch. Acta Crystallogr D Biol Crystallogr. 2009;D65:875–879. doi: 10.1107/S0907444909018800. [DOI] [PubMed] [Google Scholar]
- Fernandez-Fuentes N, Hermoso A, Espandaler J, Querol E, Aviles FX, Oliva B. Classification of common functional loops of kinase superfamilies. Proteins: Structure, function and bioinformatics. 2004;56:539–555. doi: 10.1002/prot.20136. [DOI] [PubMed] [Google Scholar]
- Guedin A, Lacroix L, Mergny JL. Thermal melting studies of ligand-DNA interactions. Methods Mol Biol. 2010;613:25–35. doi: 10.1007/978-1-60327-418-0_2. [DOI] [PubMed] [Google Scholar]
- Hayes F, Van Melderen L. Toxin-antitoxins: diversity, evolution and function. Crit Rev Biochem Mol Biol. 2011;46:386–408. doi: 10.3109/10409238.2011.600437. [DOI] [PubMed] [Google Scholar]
- Huse M, Kuriyan J. The conformational plasticity of protein kinases. Cell. 2004;109:275–282. doi: 10.1016/s0092-8674(02)00741-9. [DOI] [PubMed] [Google Scholar]
- Johnson LN, Lewis RJ. Structural basis for control by phosphorylation. Chem Rev. 2001;101:2209–2242. doi: 10.1021/cr000225s. [DOI] [PubMed] [Google Scholar]
- Jones TA, Zou J-Y, Cowan SW, Kjeldgarrd M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. 1991;A47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- Korch SB, Henderson TA, Hill TM. Characterization of the hipA7 allele of Escherichia coli and evidence that high persistence is governed by (p)ppGpp synthesis. Molec Microbiol. 2003;50:1199–1213. doi: 10.1046/j.1365-2958.2003.03779.x. [DOI] [PubMed] [Google Scholar]
- Korch SB, Hill TM. Ectopic overexpression of wild-type and mutant hipA genes in Escherichia coli: effects on macromolecular synthesis and persister formation. J Bacteriol. 2006;188:3826–3836. doi: 10.1128/JB.01740-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lãzar V, Chifiriuc MC. Medical significance and new therapeutical strategies for biofilm associated infections. Roum Arch Microbiol Immunol. 2010;69:125–138. [PubMed] [Google Scholar]
- Lewis K. Persister cells, dormancy and infectious disease. Nat Rev Microbiol. 2007;5:48–56. doi: 10.1038/nrmicro1557. [DOI] [PubMed] [Google Scholar]
- Lewis K. Persister cells. Ann Rev Microbiol. 2010;6:357–372. doi: 10.1146/annurev.micro.112408.134306. [DOI] [PubMed] [Google Scholar]
- Moyed HS, Broderick SH. Molecular cloning and expression of hipA, a gene of Escherichia coli K-12 that affects frequency of persistence after inhibition of murein synthesis. J Bacteriol. 1986;166:399–403. doi: 10.1128/jb.166.2.399-403.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moyed HS, Bertrand KP. hipA, a newly recognized gene of Escherichia coli K-12 that affects frequency of persistence after inhibition of murein synthesis. J Bacteriol. 1983;155:768–775. doi: 10.1128/jb.155.2.768-775.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira SFF, Goss L, Dworkin J. Eukaryotic-like serine/threonine kinases and phosphatases in bacteria. Microbiol Mol Biol R. 2011;75:192–212. doi: 10.1128/MMBR.00042-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumacher MA, Piro KM, Xu W, Hansen S, Lewis K, Brennan RG. Molecular mechanisms of HipA-mediated multidrug tolerance and its neutralization by HipB. Science. 2009;323:396–401. doi: 10.1126/science.1163806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor SS, Kornev PA. Protein kinases: evolution of dynamic regulatory proteins. Trends Biochem Sci. 2011;36:65–77. doi: 10.1016/j.tibs.2010.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waldron TT, Murphy KP. Stabilization of proteins by ligand binding: application to drug screening and determination of unfolding energetics. Biochemistry. 2003;42:5058–5064. doi: 10.1021/bi034212v. [DOI] [PubMed] [Google Scholar]
- Welburn JPI, Tucker JA, Johnson T, Lindert L, Morgan M, Willis A, Nobel ME, Endicott JA. How tyrosine 15 phosphorylation inhibits the activity of cyclin dependent kinase 2-cyclin A. J Biol Chem. 2007;282:3173–3181. doi: 10.1074/jbc.M609151200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


