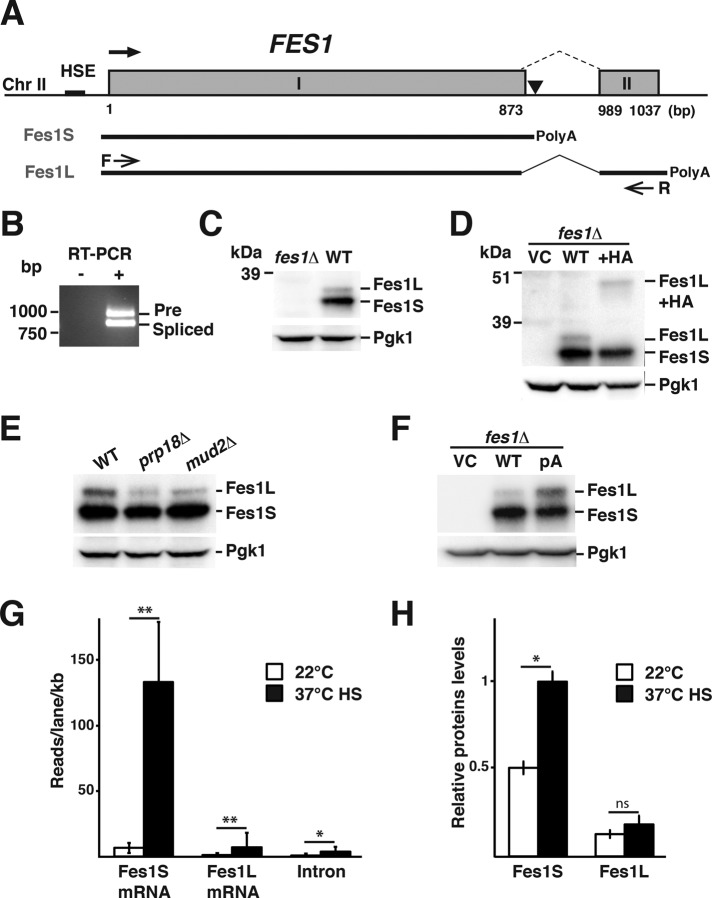
FIGURE 1:
Heat shock–regulated alternative splicing of FES1 transcripts results in the expression of Fes1L and Fes1S. (A) The heat shock element–promoted (HSE) FES1 locus contains sequence elements that encode two exons (I and II) separated by an intron (dashed line). Use of the promoter-proximal site of polyadenylation in the intron (▼) supports the expression of Fes1S from a transcript that encompasses exon I. Alternative splicing removes the promoter- proximal polyadenylation site in the intron and supports expression of Fes1L from exons I and II. (B) RT-PCR analysis of transcripts that encompass exon I and II, using primers F and R as indicated in A. The longer transcript (Pre) contains the intron, and the shorter transcript is splicing (Spliced) and supports the expression of Fes1L. (C) Western analysis of protein extracts from WT and fes1Δ with rabbit serum raised against recombinant Fes1S. Both Fes1S (32.6 kDa) and Fes1L (34.2 kDa) are expressed from the endogenous FES1 locus. Parallel blotting against Pgk1 functions as a loading control. (D) Western analysis as in C of fes1Δ cells transformed with vector control (VC) or derivatives that carry either the WT FES1 locus or a version with a 9.4- kDa 6×HA (HA) fused in-frame to the 3′ end of exon II. (E) Western analysis as in C of WT and splicing-impaired mutants prp18Δ and mud2Δ. (F) Western analysis as in C of fes1Δ cells transformed with VC or derivatives that carry either WT FES1 locus or a derivative with a T-to-C substitution mutation at the polyadenylation site indicated in A. (G) Quantification of FES1 transcript variants from Illumina 1G Analyzer reads under growth of WT yeast at 22°C in YPD medium and after transient heat shock (HS); 37°C, 15 min. Data are presented as reads per lane per kilobase for mRNA. Error bars indicate SD of data from two biological replicates. (H) Quantification of Fes1L and Fes1S expression levels using Western analysis under the same conditions as in G. Error bars indicate SD (N = 3). *p < 0.05, **p < 0.01, ns > 0.05.