Figure 1.
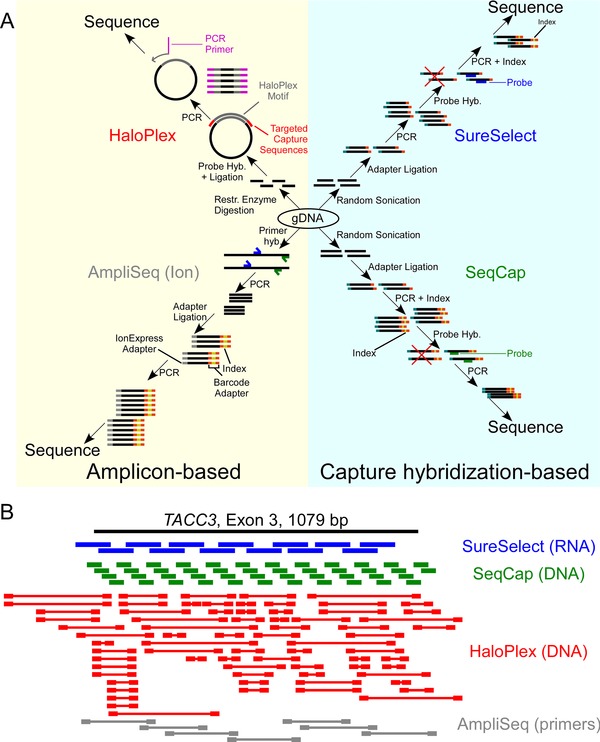
Whole exome capture methods and probe strategy. A: Schematic of experimental procedures of all four methods. SureSelect and SeqCap are classified as capture based due to their sonication‐based fragmentation method and the use of oligonucleotides to hybridize and capture target regions, whereas HaloPlex and AmpliSeq are classified as amplicon based due to their use of use of oligonucleotides as PCR primers for amplicons. B: Probe or amplicon lay out targeting a particular target exon (TACC3, exon 3, chr4:1729435–1730514). The probe layout of SeqCap, in this diagram, is an approximation, since these probe coordinates are not publicly available. SeqCap and SureSelect utilize staggered probes, and Ampliseq uses adjacent amplicons, whereas HaloPlex's probes are complimentary only to sequences near ends of the amplicons, and the middle of the probe is a DNA motif used for HaloPlex‐specific purposes.
