Abstract
Objective
To functionally characterize the osteoarthritis (OA) susceptibility variants that map to a region of high linkage disequilibrium (LD) on chromosome 20q13 marked by the single‐nucleotide polymorphism (SNP) rs6094710 and encompassing NCOA3 and SULF2.
Methods
Nucleic acids were extracted from the cartilage of OA patients. Overall and allelic expression of NCOA3 and SULF2 were measured by quantitative reverse transcription–polymerase chain reaction and pyrosequencing, respectively. The functional effect of SNPs within the 20q13 locus was assessed in vitro using luciferase reporter constructs and electrophoretic mobility shift assays (EMSAs). The in vivo effect of nuclear receptor coactivator 3 (NCOA3) protein depletion on primary human OA articular cartilage chondrocytes was assessed using RNA interference.
Results
Expression of NCOA3 correlated with the genotype at rs6094710 (P = 0.006), and the gene demonstrated allelic expression imbalance (AEI) in individuals heterozygous for the SNP (mean AEI 1.21; P < 0.0001). In both instances, expression of the OA‐associated allele was reduced. In addition, there was reduced enhancer activity of the OA‐associated allele of rs116855380, a SNP in perfect LD with rs6094710 in luciferase assays (P < 0.001). EMSAs demonstrated a protein complex binding with reduced affinity to this allele. Depletion of NCOA3 led to significant changes (all P < 0.05) in the expression of genes involved in cartilage homeostasis.
Conclusion
NCOA3 is subject to a cis‐acting expression quantitative trait locus in articular cartilage, which correlates with the OA association signal and with the OA‐associated allele of the functional SNP rs116855380, a SNP that is located only 10.3 kb upstream of NCOA3. These findings elucidate the effect of the association of the 20q13 region on OA cartilage and provide compelling evidence of a potentially causal candidate SNP.
Osteoarthritis (OA) is a common, multifactorial disease characterized by progressive focal loss of articular cartilage, which can be accompanied by changes in the function of other tissues in the joint 1. Genetics is a major risk factor for OA and acts via a large number of susceptibility alleles of small individual effect sizes 2.
Several recent genome‐wide association studies (GWAS) have identified susceptibility loci for OA 3, 4, 5, including a recent meta‐analysis of >78,000 European subjects in whom a genome‐wide significant signal associated with hip OA was identified 6. This signal, at chromosome 20q13, was marked by the single‐nucleotide polymorphism (SNP) rs6094710 (G/A) (P = 7.9 × 10− 9; odds ratio 1.28). This SNP is moderately rare, with a minor allele frequency (MAF) of 0.04 (4%) in Europeans. It is located upstream of NCOA3 and SULF2, genes that encode nuclear receptor coactivator 3 and extracellular heparan sulfate 6‐O‐endosulfatase 2, respectively. Expression of NCOA3 is reduced in OA cartilage compared to preserved cartilage from the same joint 6, whereas SULF2 expression is increased in OA cartilage compared to normal articular cartilage 7.
The SNP rs6094710 is in perfect linkage disequilibrium (LD) (r2 = 1, D′ = 1) with 11 other SNPs within the 20q13 region. There are no other SNPs with an r2 value of >0.6 with these 12 SNPs. This strongly implies that the association signal is mediated by 1 of the 12 SNPs, all of which encompass a region of 194 kb that includes all of NCOA3 and approximately two‐thirds of SULF2. One of the 12 SNPs, rs6094752, is a missense SNP leading to an amino acid change (Arg>Cys) at position 218 in the NCOA3 protein 6. However, this substitution is predicted to be benign, according to a study in which prediction tools were used 8.
The vast majority of risk alleles for common diseases modulate susceptibility by acting as expression quantitative trait loci (eQTLs), which influence the expression or stability of a transcript 9, 10, 11. In OA, an excellent example is rs143383, which is located in the 5 ′‐untranslated region of GDF5; the T allele of this SNP correlates with reduced GDF5 expression in the joint tissue of OA patients 12. We hypothesized, therefore, that the association with OA susceptibility marked by rs6094710 is an eQTL that acts on NCOA3 and/or SULF2, and the functionality of this eQTL can be directly linked to either rs6094710 or 1 of the 11 SNPs that are in perfect LD with it. We tested this hypothesis by examining both overall gene expression and allelic expression of NCOA3 and SULF2 in cartilage tissue from OA patients, and then used a number of experimental methods to further pursue the identified functionality.
PATIENTS AND METHODS
Patients
The Newcastle and North Tyneside research ethics committee (REC) granted ethics approval (REC reference no. 09/H0906/72) to obtain cartilage tissue from patients with primary OA undergoing elective hip or knee replacement. Informed consent was obtained from each donor. Full‐thickness, macroscopically normal articular cartilage away from the OA lesion was collected. Further information regarding the 65 patients is provided in Supplementary Table 1 (available on the Arthritis & Rheumatology web site at http://onlinelibrary.wiley.com/doi/10.1002/art.39278/abstract). Nucleic acids were extracted and complementary DNA (cDNA) was synthesized as previously described 13, 14.
Evaluation of gene expression
Gene expression was assessed by quantitative real‐time reverse transcription–polymerase chain reaction (RT‐PCR) using an ABI Prism 7900HT Sequence Detection System (Applied Biosystems). NCOA3 reactions were performed using a TaqMan Gene Expression Assay (Hs01105251_m1; Applied Biosystems). All other reactions were performed using PrimeTime quantitative PCR assays (Integrated DNA Technologies). A list of the primer and probe sequences used for RT‐PCR is available in Supplementary Table 2 (available on the Arthritis & Rheumatology web site at http://onlinelibrary.wiley.com/doi/10.1002/art.39278/abstract). For each cDNA sample, 3 pipetting replicates were performed to determine the expression levels of each gene. These expression levels, relative to those of the housekeeping genes 18S, GAPDH, and HPRT1, were calculated using the formula . Outliers were removed from the data using Grubbs’ test 15. P values were calculated using a Mann‐Whitney 2‐tailed exact test.
Transcript SNP selection and allelic expression analysis
The SNP rs6094752 was used for NCOA3, since this transcript SNP is in perfect LD with rs6094710. It is located in exon 7 of the gene. The SNP rs3810526 was used for SULF2, since this transcript SNP has the highest pairwise LD with rs6094710 (r2 = 0.006, D′ = 0.40). The SNP rs3810526 is a G/A amino acid transition, has an MAF of 0.34 in Europeans, and is located in exon 3 of SULF2.
Allelic expression imbalance (AEI) was determined as previously described 16. Standard PCR, using a G‐Storm GS4 Q4 Quad Block Thermal Cycler (Somerton Biotechnology Centre), was used to amplify the rs6094752 and rs3810526 regions with biotinylated primers, creating biotinylated PCR products. Samples were then analyzed on a PyroMark Q24 MDx platform (Qiagen) using a PyroMark Gold Q96 Reagents kit, in accordance with the manufacturer's instructions. The primers used for each SNP, including the sequencing primers, were obtained from Sigma‐Aldrich (a list is provided in Supplementary Table 2, available on the Arthritis & Rheumatology web site at http://onlinelibrary.wiley.com/doi/10.1002/art.39278/abstract) and were designed to reside within single exons. Sequences were generated automatically, and an output of allelic ratio was produced using PSQ 96 SQA software (Qiagen).
For each patient, samples were analyzed in triplicate, cartilage cDNA and cartilage DNA were analyzed concurrently, and allelic expression of cDNA was normalized to that of its corresponding DNA. Accurate discrimination of SNP alleles was verified using artificially created allelic expression ratios derived from DNAs of known genotype. P values for each SNP were calculated by comparing DNA allelic expression ratios to cDNA allelic expression ratios. Comparisons were made using a Mann‐Whitney 2‐tailed exact test. In addition, pyrosequencing was used to genotype rs6094710.
Construction of luciferase reporter plasmids, cell cultures, and luciferase assays
Two online databases were searched to identify SNPs within the 20q13 region that are in LD with rs6094710: the Broad SNAP database, which encompasses data from the 1000 Genomes project (http://www.broadinstitute.org/mpg/snap/), and the HapMap database (http://hapmap.ncbi.nlm.nih.gov). To generate the constructs for use in the luciferase reporter gene assay, primers containing either an Mlu I or an Xho I restriction enzyme site toward their 5 ′ ends were designed to amplify DNA regions encompassing each of the 12 SNPs. Primer sequences are listed in Supplementary Table 3 (available on the Arthritis & Rheumatology web site at http://onlinelibrary.wiley.com/doi/10.1002/art.39278/abstract). Template DNA consisted of a mix of DNA from 4 patients heterozygous for rs6094710.
PCR products were purified using a QIAquick PCR purification kit (Qiagen) and cloned into the pGL3‐promoter luciferase reporter plasmid (Promega) using the Mlu I and Xho I restriction sites. Positive clones were sequenced to ensure the correct sequence of the constructs. The SW1353 human chondrosarcoma cell line and the SW872 human liposarcoma cell line (both from ATCC) were transfected for the luciferase assays. Twenty‐four hours before transfection, SW1353 cells were seeded at a density of 6,000 per well, while SW872 cells were seeded at a density of 10,000 per well, in a 96‐well plate. Cells were transfected with 50 ng of pGL3 construct DNA and 30 ng Renilla luciferase reporter vector pRL‐TK (Promega), using FuGene HD transfection reagent (Promega). After 24 hours, the cells were lysed and luciferase activity was determined using a Dual‐Luciferase Reporter Assay System (Promega). Luminescence was measured using a GloMax‐Multi Detection System (Promega). Firefly luciferase activity was normalized to the activity of Renilla luciferase. Six technical and 6 biologic repeats were performed per cell line per construct. P values were calculated using a Mann‐Whitney 2‐tailed exact test.
Electrophoretic mobility shift assays (EMSAs)
The JASPAR, PROMO, and TFSearch online databases were used to predict protein binding to the A and G alleles of rs116855380 17, 18, 19. Nuclear protein was extracted from SW1353 and SW872 cells as previously described 20. Forward and reverse single‐stranded DY682‐labeled oligonucleotides (Eurofins MWG Operon) for both alleles, spanning 15 bp to each side of rs116855380, were annealed to generate double‐stranded probes, and EMSAs were performed as previously described 20.
All EMSA experiments were performed twice. Briefly, binding reactions were performed for 20 minutes at room temperature using an Odyssey EMSA buffer kit (LiCor Biosciences). Each reaction contained 5 μg nuclear extract, 200 fmoles annealed oligonucleotide, 1 × Binding Buffer, 2.5 mM dithiothreitol, 0.5 μg poly(dI‐dC), and 10 mM EDTA. Samples were separated on a 5% (weight/volume) native polyacrylamide gel in 0.5 × Tris–borate–EDTA for 4 hours at 100 volts, followed by visualization with an Odyssey Infrared Imager (LiCor Biosciences). For competition assays, unlabeled oligonucleotides identical to the probes, or containing a transcription factor consensus sequence, were added to the binding reaction in excess. For supershift assays, 2 μg or 6 μg of antibody was added to the binding reaction. All probe and competitor sequences and antibodies used in the EMSAs are listed in Supplementary Tables 4 and 5 (available on the Arthritis & Rheumatology web site at http://onlinelibrary.wiley.com/doi/10.1002/art.39278/abstract).
Immunohistochemistry
Macroscopically normal cartilage away from the OA lesion was fixed overnight in 10% formalin (Sigma‐Aldrich) and wax embedded. Sections were cut and stained using anti‐NCOA3 antibody (ab133611; Abcam). Briefly, sections were incubated in boiling citrate, pH 6.0, for 6 minutes, washed in Tris buffered saline for 5 minutes, and stained using an ImmPRESS Anti‐Rabbit Ig Reagent Kit (Vector Laboratories), in accordance with the manufacturer's instructions. Primary antibody was applied for 30 minutes at a 1:150 dilution, diaminobenzidine was used as the peroxidase substrate, and hematoxylin was used to counterstain.
RNA interference
Primary human articular chondrocytes (HACs) were isolated by enzymatic digestion of OA cartilage and cultured as described previously 21. HACs were seeded 24 hours before transfection with 100 nM Dharmacon ON‐TARGETplus SMARTpool small interfering RNA (siRNA) targeted against NCOA3 (L‐003759‐00) or an ON‐TARGETplus nontargeting siRNA control pool (D‐001810‐10‐20), using DharmaFECT 1 transfection reagent (Dharmacon). After 48 hours, gene expression was assessed by RT‐PCR, and protein levels were assessed by immunoblotting. For RT‐PCR, cells were seeded at a density of 10,000 cells per well in a 96‐well plate, with 6 technical repeats per treatment. Forty‐eight hours after transfection, the 96‐well plate was used to synthesize cDNA using a Cells‐to‐cDNA II kit (Ambion), and gene expression was assessed by RT‐PCR.
For immunoblot analysis, cells were seeded at a density of 350,000 cells per well in a 6‐well plate. Two wells were seeded per treatment and their contents combined before protein extraction. Total protein was extracted from the 6‐well plate using a spin column extraction kit (Nucleospin RNA/Protein; Macherey‐Nagel, supplied by Fisher), in accordance with the manufacturer's instructions. Three biologic repeats were performed using cells obtained from 3 individual OA patients. P values were calculated using a Student's 2‐tailed t‐test.
Immunoblotting
To assess depletion of NCOA3, total protein from HACs was quantified (Bradford reagent; Expedeon) and 10 μg was resolved on 12% (w/v) sodium dodecyl sulfate–polyacrylamide gels. Protein was transferred to Immobilon‐P PVDF membranes (Merck Millipore). Anti‐NCOA3 antibody (ab133611; Abcam) was used at a 1:1,000 dilution to assess protein levels. A β‐actin antibody (A5316; Sigma) was used at a 1:10,000 dilution as a loading control. Depletion of NCOA3 was quantified using ImageJ software 22, and NCOA3 levels were normalized to the levels of β‐actin for each treatment condition.
RESULTS
Quantitative expression of NCOA3 and SULF2 in OA cartilage stratified by rs6094710 genotype
To determine whether the 20q13 locus harbors a cartilage eQTL that correlates with the OA association signal, we quantified the expression levels of NCOA3 and SULF2 in articular cartilage samples from patients with OA and stratified the data by genotype at rs6094710. Although rs6094710 is associated only with hip OA, we performed our analyses in both hip and knee tissue in order to determine whether the functional variant operates in knee as well as hip tissue. Since this SNP has a moderately low MAF (0.04), none of our patients were homozygous for the minor allele of the SNP. Our comparisons therefore involved an analysis of GG homozygotes versus GA heterozygotes.
We observed a significant correlation between NCOA3 expression and genotype (Figure 1A), with presence of the minor A allele of the SNP correlating with reduced gene expression (P = 0.006). The A allele of rs6094710 has been found to be more common in OA patients compared to controls 6.
Figure 1.
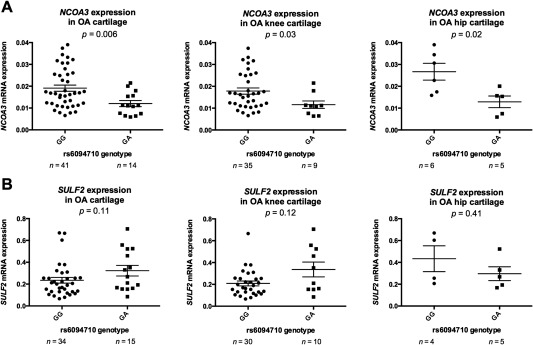
Columnar scatter plots of the quantitative expression of NCOA3 (A) and SULF2 (B) in osteoarthritis (OA) cartilage, stratified by genotype at the OA‐associated single‐nucleotide polymorphism rs6094710. In cartilage samples from all OA patients (left), patients with knee OA (center), and patients with hip OA (right), the expression of each gene was assessed by quantitative real‐time reverse transcription–polymerase chain reaction, with values normalized to the values for the housekeeping genes 18S, GAPDH, and HPRT1. Individual symbols represent the average of the 3 replicates for each sample; horizontal lines represent the mean ± SEM. P values were calculated by Mann‐Whitney 2‐tailed exact test.
We next separated the OA patients into those with knee OA (Figure 1A) and those with hip OA (Figure 1A). We observed a significant correlation between NCOA3 expression and genotype both in patients with knee OA and in patients with hip OA (P = 0.03 and P = 0.02, respectively).
Finally, we stratified NCOA3 expression by joint site, by sex, and by age at the time of joint replacement. We found no correlations between the level of NCOA3 expression and any of these 3 parameters (all P > 0.05; results in Supplementary Figure 1, available on the Arthritis & Rheumatology web site at http://onlinelibrary.wiley.com/doi/10.1002/art.39278/abstract).
There was no correlation between rs6094710 genotype and SULF2 expression (P = 0.11) (Figure 1B). We repeated the analysis after separating patients into those with knee OA (Figure 1B) and those with hip OA (Figure 1B). Again, there was no correlation between rs6094710 genotype and SULF2 expression in patients with knee OA or those with hip OA (P = 0.12 and P = 0.41, respectively). When we stratified SULF2 expression by joint site, by sex, and by age at the time of joint replacement, we found that there was greater gene expression in the hip than in the knee (P = 0.04) and there was increased expression of SULF2 in women compared to men (P = 0.04), but there was no correlation with age (P = 0.33) (results in Supplementary Figure 1, available on the Arthritis & Rheumatology web site at http://onlinelibrary.wiley.com/doi/10.1002/art.39278/abstract).
Overall, our analysis revealed an eQTL operating on NCOA3 that correlated with the OA association signal. Moreover, we found that the OA risk allele was mediating the reduced expression of the gene.
AEI analysis of NCOA3 and SULF2 in OA cartilage
We further investigated the eQTL by AEI analysis, which measures the messenger RNA (mRNA) output from each allele of a SNP in heterozygous individuals. For NCOA3, we used transcript SNP rs6094752, which is in perfect LD (r2 = 1, D′ = 1) with the OA‐associated SNP rs6094710. We genotyped 130 patients for whom DNA and RNA samples were available, and identified 8 individuals heterozygous for rs6094710. For SULF2, we chose the transcript SNP rs3810526, which has the highest pairwise LD with rs6094710. This LD is nevertheless low (r2 = 0.006, D′ = 0.40), and this, along with the low MAF of rs6094710, meant that we were able to identify only 3 compound heterozygous patients among our cohort.
Cartilage from 6 patients with knee OA and 2 patients with hip OA was analyzed for AEI at rs6094752, with the T allele (equivalent to the rs6094710 A allele) showing reduced expression in 7 patients (Figure 2). The mean fold difference in allelic expression was 1.21, indicating that the C allele of the SNP correlated with 21% more NCOA3 expression than the OA‐associated T allele (P < 0.0001).
Figure 2.
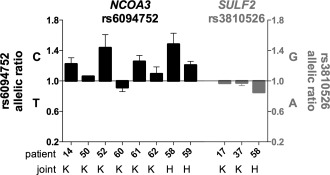
Allelic expression analysis in cartilage samples from osteoarthritis (OA) patients. Allelic expression was assessed using the transcript single‐nucleotide polymorphisms rs6094752 in NCOA3 (black bars) and rs3810526 in SULF2 (gray‐shaded bars). Allelic expression of cDNA was normalized to that of its corresponding DNA. Data are presented as a ratio of expression of the major allele over that of the minor allele; thus, a value above 1 means that there is more of the major allele. Samples are grouped according to OA joint site (knee [K] or hip [H]), with the numbers of patients shown. Bars show the mean ± SEM.
Cartilage from 2 patients with knee OA and 1 patient with hip OA was analyzed for AEI at rs3810526. One of the 3 patients (patient 58) showed increased expression of the A allele (Figure 2). The mean fold difference in allelic expression was 0.93, indicating that the A allele of the SNP produced 7% more SULF2 than the G allele (P = 0.04).
Overall, the AEI data confirmed that the risk allele of the OA association signal correlated with reduced expression of NCOA3.
Identifying rs116855380 as a functional SNP
As already noted, rs6094710 is in perfect LD with 11 SNPs within the 20q13 OA locus (for details regarding the physical location of the SNPs, see Table 1; a visual representation of the locus can be found in Supplementary Figure 2, available on the Arthritis & Rheumatology web site at http://onlinelibrary.wiley.com/doi/10.1002/art.39278/abstract). There are no other SNPs within the region showing LD at an r2 value of >0.6. To assess whether rs6094710 or any of the other 11 SNPs are functional, we cloned the regions of DNA containing each SNP separately into a luciferase reporter plasmid. We created both allelic forms for each SNP and performed reporter gene expression assays using the SW1353 and SW872 cell lines.
Table 1.
Luciferase gene reporter assays in SW1353 and SW872 cell lines of the single‐nucleotide polymorphisms (SNPs) in perfect linkage disequilibrium with rs6094710*
| SNP, allele (major, minor) | SNP location | Predicted enhancer | SW1353 cells | SW872 cells | ||||
|---|---|---|---|---|---|---|---|---|
| Luciferase activity | Fold difference (major/minor) | P | Luciferase activity | Fold difference (major/minor) | P | |||
| rs6094710 | ||||||||
| G | Intergenic | Yes | 3.8 | 1.1 | 0.79 | 5.3 | 1.0 | 0.87 |
| A | 3.5 | 5.1 | ||||||
| rs73122077 | ||||||||
| C | Intergenic | Yes | 1.0 | 1.0 | 0.32 | 1.4 | 1.1 | 0.015 |
| T | 1.0 | 1.3 | ||||||
| rs6090683 | ||||||||
| G | Intergenic | Yes | 1.3 | 1.1 | 0.67 | 1.4 | 1.2 | 0.069 |
| C | 1.2 | 1.2 | ||||||
| rs6090684 | Intergenic | Yes | 1.3 | 0.013 | 0.9 | 0.043 | ||
| G | 0.9 | 0.9 | ||||||
| A | 0.7 | 1.0 | ||||||
| rs116855380 | ||||||||
| A | Intergenic | Yes | 1.4 | 1.2 | 0.0003 | 1.6 | 1.5 | <0.0001 |
| G | 1.2 | 1.1 | ||||||
| rs117212926 | ||||||||
| G | Intronic (NCOA3) | No | 0.6 | 0.9 | 0.53 | 1.3 | 0.9 | 0.71 |
| A | 0.7 | 1.5 | ||||||
| rs72662711 | ||||||||
| T | Intronic (NCOA3) | Yes | 1.1 | 1.0 | 0.99 | 2.0 | 1.5 | 0.0014 |
| C | 1.0 | 1.3 | ||||||
| rs6090704 | ||||||||
| G | Intronic (NCOA3) | Yes | 1.0 | 0.9 | 0.61 | 1.6 | 1.2 | 0.027 |
| A | 1.1 | 1.3 | ||||||
| rs6094752 | ||||||||
| C | Exonic (NCOA3) | No | 0.8 | 1.1 | 0.70 | 2.0 | 1.1 | 0.33 |
| T | 0.8 | 1.8 | ||||||
| rs72645259 | ||||||||
| T | Intronic (NCOA3) | No | 2.3 | 0.9 | 0.34 | 2.6 | 0.7 | 0.0021 |
| C | 2.6 | 4.0 | ||||||
| rs73913405 | ||||||||
| C | Intronic (SULF2) | Yes | 0.7 | 1.0 | 0.15 | 1.3 | 1.0 | 0.76 |
| T | 0.6 | 1.3 | ||||||
| rs73913406 | ||||||||
| T | Intronic (SULF2) | Yes | 1.5 | 1.5 | <0.0001 | 1.2 | 1.2 | 0.13 |
| G | 1.0 | 1.0 | ||||||
Luciferase activity was calculated as the activity of firefly luciferase relative to Renilla luciferase, with results normalized to the values in empty vector. P values were calculated by Mann‐Whitney 2‐tailed exact test.
In SW1353 cells, we observed significant allelic differences (P < 0.05) in luciferase activity for 3 SNPs, while in SW872 cells, we observed significant differences in luciferase activity between the alleles of 6 SNPs (Table 1). Of all the SNPs tested, only 2 of the SNPs, rs116855380 and rs6090684, showed significant differences in both cell lines. However, the allelic differences observed for rs6090684 acted in opposite directions between the 2 cell lines.
The region of DNA containing rs116855380 displayed enhancer activity in both cell lines when the A allele of the SNP was present, and the presence of the G allele was associated with a significant decrease in reporter activity in SW1353 cells (P = 0.0003) (Figure 3A) and SW872 cells (P < 0.0001) (Figure 3B). The G allele of rs116855380 is in perfect LD with the OA‐associated A allele of rs6094710; thus, these findings from luciferase reporter assays supported the findings from analyses of NCOA3 expression and AEI.
Figure 3.
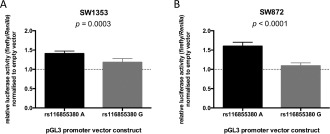
Results of luciferase reporter assays for the major A allele and minor G allele of rs116855380 in SW1353 cells (A) and SW872 cells (B), assessing firefly luciferase activity relative to Renilla luciferase activity, normalized to an empty pGL3‐promoter vector (indicated by the broken line). Bars show the mean ± SEM fold change from 6 biologic repeats, each with 6 technical replicates. P values were calculated by Mann‐Whitney 2‐tailed exact test.
EMSA assessment of trans‐acting factors binding to rs116855380
An analysis of ENCODE data sets (http://genome.ucsc.edu/ENCODE/) revealed that rs116855380 is predicted to lie within a region of enhancer activity 23, a prediction that was supported by the findings from our luciferase assays. We therefore investigated protein complex binding to rs116855380 using nuclear protein extracted from SW1353 and SW872 cells and using fluorescence‐labeled A and G allele probes representing the 2 alleles of this SNP (results in SW1353 cells shown in Figure 4A; results in SW872 cells shown in Supplementary Figure 3, available on the Arthritis & Rheumatology web site at http://onlinelibrary.wiley.com/doi/10.1002/art.39278/abstract).
Figure 4.
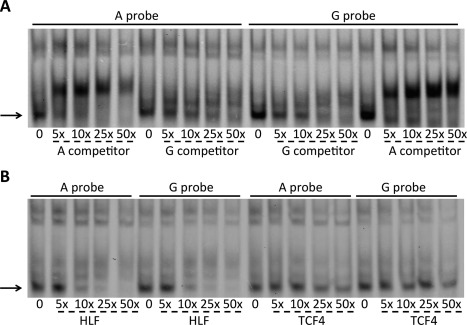
Electrophoretic mobility shift assay (EMSA) analyses in SW1353 cells. A, Increasing concentrations of unlabeled A and G allele competitor were added to the EMSA reaction containing cell nuclear extract and the A and G allele probes. Arrow indicates the specific complex binding to the probes. B, Increasing concentrations of hepatic leukemia factor (HLF) and transcription factor 4 (TCF‐4) unlabeled consensus competitors were added to the EMSA reaction containing the A or G allele probe. Arrow indicates the complex that is competed.
We observed a similar pattern of protein complex binding to the 2 probes in SW1353 cells. We tested the specificity of the assay by adding unlabeled A and G allele competitors, and found that one complex bound specifically to the rs116855380 probes (Figure 4A). Binding of this complex to both probes was outcompeted with a lower concentration of the A allele competitor than that of the G allele competitor. For example, Figure 4A shows that a lower concentration of the A allele competitor, as compared to the G allele competitor, was required to outcompete the A allele probe. The addition of the A allele competitor at 25× and 50× concentrations resulted in the complete disappearance of the complex (Figure 4A), whereas these same concentrations of the G allele competitor resulted in only a reduction in the intensity of these bands (Figure 4A). The same effect could be observed in SW872 cells and in repeated experiments (results in Supplementary Figure 3, available on the Arthritis & Rheumatology web site at http://onlinelibrary.wiley.com/doi/10.1002/art.39278/abstract).
Using the online databases JASPAR, PROMO, and TFSearch, we identified 5 transcription factors (transcription factor 4 [TCF‐4], lymphoid enhancer–binding factor 1 [LEF‐1], hepatic leukemia factor [HLF], serum response factor [SRF], and myocyte enhancer factor 2C [MEF‐2C]) predicted to bind to the region containing rs116855380. We refined the number of potential factors using competitors containing the consensus binding sequence of each factor (the list of competitor sequences is provided in Supplementary Table 4, available on the Arthritis & Rheumatology web site at http://onlinelibrary.wiley.com/doi/10.1002/art.39278/abstract). With the addition of an HLF competitor, complex binding to both probes was reduced, and this effect was observed with nuclear extract from both cell lines (results in SW1353 cells shown in Figure 4B; results in SW872 cells shown in Supplementary Figure 3, available on the Arthritis & Rheumatology website at http://onlinelibrary.wiley.com/doi/10.1002/art.39278/abstract). In experiments using SW1353 cell nuclear extract, when a TCF‐4 competitor was added, complex binding to both probes was competed (Figure 4B). Little or no competition was observed when the TCF‐4 competitor was used with SW872 cell extract.
Addition of antibodies targeting TCF‐4 or HLF did not result in a supershift of the specific complex (data not shown). Through literature searches, we identified 2 transcription factors closely related to HLF (DNA binding protein and CCAAT/enhancer binding protein) that have very similar consensus binding sites 24. However, addition of antibodies to these proteins also did not result in supershift of the complex (data not shown).
Regulation of COL2A1, RUNX2, and MMP13 transcriptional activity by NCOA3
To assess the presence and distribution of NCOA3 protein, we stained cartilage tissue prepared from nonlesional OA cartilage with an anti‐NCOA3 antibody (details shown in Supplementary Figure 4, available on the Arthritis & Rheumatology web site at http://onlinelibrary.wiley.com/doi/10.1002/art.39278/abstract). The protein is present in chondrocytes in both knee and hip cartilage. Cells within the superficial and middle zones stained more strongly than cells in the deep zone. Staining was nuclear, consistent with the role of NCOA3 as a coactivator of various nuclear receptors 25, 26.
To investigate the role of NCOA3 in maintaining healthy cartilage, we analyzed the effect of NCOA3 depletion by RNA interference in HACs isolated from nonlesional areas of the OA cartilage. The depletion of NCOA3 mRNA (75% reduction) was confirmed by RT‐PCR, and the depletion of NCOA3 protein (76% reduction) was confirmed by immunoblotting (Figures 5A and B). We assessed the levels of 5 chondroprotective/anabolic genes (COL1A1, COL2A1, ACAN, SOX9, and TIMP1) and 4 genes involved in cartilage hypertrophy/catabolism (RUNX2, ADAMTS5, MMP1, and MMP13). Expression of COL2A1 was significantly increased following NCOA3 depletion (P = 0.02) (Figure 5C), whereas expression of MMP13 and RUNX2 was significantly decreased (P = 0.03 and P = 0.0002, respectively) (Figure 5C).
Figure 5.
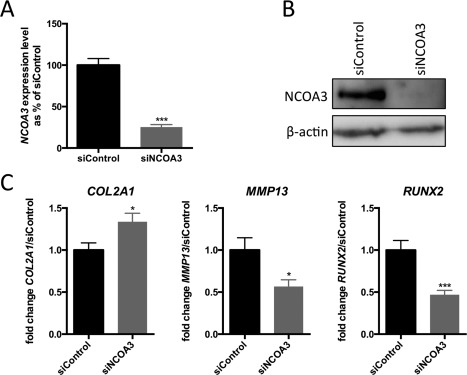
Effects of NCOA3 depletion on expression of genes involved in cartilage homeostasis. Knockdown of NCOA3 was performed in primary human articular chondrocytes obtained from 3 patients with osteoarthritis. A, Expression levels of NCOA3 mRNA in cells following NCOA3 knockdown with small interfering RNA (siRNA) targeting NCOA3 (siNCOA3). Values are the mean and SEM percentage change in mRNA expression relative to that with the control nontargeting siRNA (siControl) (set at 100%). B, Representative immunoblot demonstrating NCOA3 depletion following siNCOA3 treatment; β‐actin was used as a loading control. C, Fold change in expression of COL2A1, MMP13, and RUNX2 following siNCOA3 treatment. Values are the mean ± SEM fold change relative to that with the siControl (set at 1.0), from 3 biologic repeats, each with 6 technical replicates. P values were calculated by Student's 2‐tailed t‐test. ∗ = P < 0.05; ∗∗∗ = P < 0.001 versus siControl.
DISCUSSION
In this study, we demonstrated the presence of a cartilage eQTL operating on NCOA3 that correlates with the OA association signal marked by SNP rs6094710. The OA risk allele, the A allele of the SNP, was associated with reduced expression of the gene. We then uncovered allelic functionality at rs116855380, a SNP in perfect LD with rs6094710; the G allele of rs116855380 segregated on the same molecule as the A allele of rs6094710 in the haplotype that harbors these 2 SNPs. Knockdown of NCOA3 highlighted the important functional role of this protein in chondrocyte biology. As far as we are aware, this is the first study in which an OA association signal derived from a GWAS has been characterized to the point at which the functional effect has been elucidated and a compelling causal candidate SNP identified.
Although rs6094710 has been demonstrated to be associated with OA of the hip, we found that the eQTL operates in both hip and knee cartilage. Many OA risk loci show joint‐specific genetic effects 2, highlighting the complex nature of OA susceptibility and the fact that pathophysiologic causes are not uniform across skeletal sites 27. Presumably, lowered NCOA3 levels have a markedly detrimental effect in the hip but not in the knee.
The OA risk marked by rs6094710 did not correlate with differential expression of SULF2. We did observe a small degree of AEI in 1 of our 3 compound heterozygous patients. We cannot prove a negative, and it may be that there is an eQTL operating on SULF2. There is, however, no compelling evidence in our study to indicate that altered expression of this gene correlates with the OA association signal.
The SNP rs116855380, which we discovered as the likely mediator of the NCOA3 eQTL, is located only 10.3 kb upstream of the gene, in a region that, according to ENCODE, has a transcriptional regulatory function. We investigated trans‐acting factors that may bind differentially to the 2 alleles of rs116855380, and which could therefore be involved in mediating the allelic expression imbalance. In EMSA analysis, we showed that the consensus binding sites for TCF‐4 and HLF competed with the rs116855380 probe for binding of a specific complex, which bound with more affinity to the A allele of the SNP. We were, however, unable to observe a supershift. This suggests that another protein with a similar consensus sequence is binding to rs116855380 and promoting transcriptional activation, and that this binding, and therefore activation, is strongest for the A allele of rs116855380.
Other SNPs within the locus showed significant differences in the reporter assays. We therefore cannot discount the possibility that one or more of these SNPs could also have functional effects in the cartilage tissue. However, and unlike rs116855380, the reporter assay effects of these other SNPs were not observed in both cell types examined or in the same direction in both cell types. Our study has therefore enabled us to prioritize rs116855380 as a SNP with consistent in vitro allelic functional effects that match the findings in our OA cartilage tissue.
In our analysis of primary HACs in monolayer, a reduction in NCOA3 protein, which mimics the genetic effect identified, resulted in an increase in COL2A1 expression and a decrease in RUNX2 and MMP13 expression. In this model system, NCOA3 is therefore functional. However, these results are paradoxical when we consider that reduced expression of NCOA3 is associated with OA risk. Although COL2A1 expression has been shown to increase in OA cartilage, probably as a compensatory mechanism 28, 29, the expression of RUNX2 and MMP13 has also been found to increase 30, 31, with RUNX2 directly up‐regulating MMP13 31, 32. Thus, one would expect to see an increase in RUNX2 and MMP13 expression when reducing the level of NCOA3 protein in chondrocytes.
Our results are, however, consistent with previous findings that showed that NCOA3 directly regulates transcription of MMP13 through coactivation of activator protein 1 and polyomavirus enhancer activator 3 33. In addition, as NCOA3 is a coactivator for several nuclear hormone receptors, and RUNX2 transcription is activated by parathyroid hormone 34, NCOA3 may also act in this capacity to regulate RUNX2, and therefore MMP13, expression. NCOA3 can also recruit the nuclear factors p300/CREB binding protein–associated factor (P/CAF) and CREB binding protein to create multisubunit coactivator complexes 25, and P/CAF has been shown to increase RUNX2 activity by acetylation 35. To clarify the role of NCOA3 in cartilage biology, more detailed functional analyses are clearly merited, encompassing gene overexpression and knockdown and the use of cells in monolayer and in 3‐dimensional cultures, the latter more accurately reflecting cartilage tissue.
In conclusion, we have identified a SNP and a functional effect that are likely to account for the OA association mapped to chromosome 20q13: rs116855380 and differential allelic expression in NCOA3 in OA cartilage. Further functional analyses of the SNP, the gene, and its protein are now justified to fully comprehend how this particular genetic risk increases disease susceptibility and how that susceptibility can be mitigated.
AUTHOR CONTRIBUTIONS
All authors were involved in drafting the article or revising it critically for important intellectual content, and all authors approved the final version to be published. Dr. Reynard had full access to all of the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Study conception and design. Gee, Rushton, Louglin, Reynard.
Acquisition of data. Gee, Rushton.
Analysis and interpretation of data. Gee, Rushton, Louglin, Reynard.
Supporting information
Figure S1. Scatter plots of the quantitative expression of NCOA3 (left) and SULF2 (right) in osteoarthritis (OA) cartilage. Data is stratified by (A) joint site, (B) sex and (C) age at surgery. The data points represent the average of the three replicates for each sample. The expression of each gene was assessed by quantitative real‐time reverse transcription PCR and normalized to the housekeeping genes 18S, GAPDH and HPRT1. n is the number of patients studied. The horizontal lines in the columnar scatter plots represent the mean and the standard error of the mean. The trendlines in the XY scatter plots represent linear regression of the data. p‐values for (A) and (B) were calculated using a two‐tailed Mann‐Whitney exact test. p‐values for (C) were calculated using linear regression.
Figure S2. Schematic representation of the 20q13 locus. Gene track is a screenshot from http://genome.ucsc.edu, using release hg19. The genes are represented by horizontal lines, with the arrows indicating the direction of gene transcription. The exons are represented by vertical bars, whose width is proportional to the length of the exon. The numbers above the gene tracks indicate the position within chromosome 20. The vertical lines beneath the gene tracks represent the SNPs analyzed in this study. The SNP that identified the signal in the GWAS, rs6094710, is labeled in white text on a black background, and the two transcript SNPs used for AEI, rs6094752 (NCOA3) and rs3810526 (SULF2), are labeled with bold text. Also labeled are the other SNPs in perfect LD with rs6094710.
Figure S3. Electrophoretic mobility shift assay (EMSA) analysis in SW872 (A and B) and SW1353 cells (C and D). (A and C) Increasing concentrations of unlabeled G and A allele competitor were added to the EMSA reaction containing the G and A allele probes and nuclear extract, with the arrow indicating the specific complex binding to the probes. (B and D) The addition of increasing concentrations of HLF and TCF4 unlabeled consensus competitors to the EMSA reaction containing the G or A allele probe. The arrow indicates the complex that is competed.
Figure S4. Expression of NCOA3 in cartilage tissue. (A,B,D,E) Immunohistochemical staining against NCOA3 in macroscopically normal knee (A and D) and hip (B and E) cartilage obtained from osteoarthritis patients. (C and F) Negative control staining with no primary antibody in the same hip sample as shown in B and E. The articular surface is at the top of the images. Top panels (A‐C) were taken at 5x magnification; bottom panels (D‐F) show enlarged views taken at 10x magnification. Cells exhibit nuclear staining, which is consistent with the role of NCOA3 as a co‐activator of various nuclear receptors. Enlarged regions are indicated by the black boxes in A‐C. Scale bars = 200 m.
Table S1. Table of OA patient characteristics, their genotype at rs6094710 and details of their use in quantitative real‐time reverse transcription PCR (RT‐PCR). F, female; M, male; K, knee; H, hip.
Table S2. Primer and probe sequences used for the quantitative real‐time reverse transcription PCR (RT‐PCR) of a panel of genes. The NCOA3 primers are not listed and were purchased from Applied Biosystems as an off‐the‐shelf TaqMan Gene Expression Assay. Also listed are the primers used for the pyrosequencing analysis of SNPs rs6094710, rs6094752 and rs3810526. FP, forward primer; RP, reverse primer; SP, sequencing primer; Pr, probe
Table S3. The twelve DNA fragments examined by luciferase analysis. Chromosome locations based on UCSC genome browser, release hg19. The sites in the cloning primers for the restriction enzymes MluI (ACGCGT) and XhoI (CTCGAG) are highlighted in bold.
Table S4. Electrophoretic mobility shift assay (EMSA) probes and competitors. The bold and underlined bases in the EMSA probes indicate the position of rs116855380. The series of underlined bases in the transcription factor competitors indicate the site of the consensus sequence for each factor.
Table S5. Antibodies used for supershift electrophoretic mobility shift assays
ACKNOWLEDGMENTS
We thank surgeons at the Newcastle upon Tyne Hospitals NHS Foundation Trust for providing us with access to patient tissue samples. We thank the patients for donating their tissue samples. We thank Katherine Johnson and Sharon Watson for technical support.
REFERENCES
- 1. Brandt KD, Dieppe P, Radin EL. Etiopathogenesis of osteoarthritis [review]. Rheum Dis Clin North Am 2008;34:531–59. [DOI] [PubMed] [Google Scholar]
- 2. Reynard LN, Loughlin J. Insights from human genetic studies into the pathways involved in osteoarthritis [review]. Nat Rev Rheumatol 2013;9:573–83. [DOI] [PubMed] [Google Scholar]
- 3. arcOGEN Consortium, arcOGEN Collaborators . Identification of new susceptibility loci for osteoarthritis (arcOGEN): a genome‐wide association study. Lancet 2012;380:815–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kerkhof HJ, Lories RJ, Meulenbelt I, Jonsdottir I, Valdes AM, Arp P, et al. A genome‐wide association study identifies an osteoarthritis susceptibility locus on chromosome 7q22. Arthritis Rheum 2010;62:499–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Styrkarsdottir U, Thorleifsson G, Helgadottir HT, Bomer N, Metrustry S, Bierma‐Zeinstra S, et al. Severe osteoarthritis of the hand associates with common variants within the ALDH1A2 gene and with rare variants at 1p31. Nat Genet 2014;46:498–502. [DOI] [PubMed] [Google Scholar]
- 6. Evangelou E, Kerkhof HJ, Styrkarsdottir U, Ntzani EE, Bos SD, Esko T, et al. A meta‐analysis of genome‐wide association studies identifies novel variants associated with osteoarthritis of the hip. Ann Rheum Dis 2014;73:2130–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Otsuki S, Taniguchi N, Grogan SP, D'Lima D, Kinoshita M, Lotz M. Expression of novel extracellular sulfatases Sulf‐1 and Sulf‐2 in normal and osteoarthritic articular cartilage. Arthritis Res Ther 2008;10:R61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, et al. A method and server for predicting damaging missense mutations. Nat Methods 2010;7:248–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cookson W, Liang L, Abecasis G, Moffatt M, Lathrop M. Mapping complex disease traits with global gene expression [review]. Nat Rev Genet 2009;10:184–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Montgomery SB, Dermitzakis ET. From expression QTLs to personalized transcriptomics [review]. Nat Rev Genet 2011;12:277–82. [DOI] [PubMed] [Google Scholar]
- 11. Freedman ML, Monteiro AN, Gayther SA, Coetzee GA, Risch A, Plass C, et al. Principles for the post‐GWAS functional characterization of cancer risk loci [review]. Nat Genet 2011;43:513–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Egli RJ, Southam L, Wilkins JM, Lorenzen I, Pombo‐Suarez M, Gonzalez A, et al. Functional analysis of the osteoarthritis susceptibility–associated GDF5 regulatory polymorphism. Arthritis Rheum 2009;60:2055–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wilkins JM, Southam L, Price AJ, Mustafa Z, Carr A, Loughlin J. Extreme context specificity in differential allelic expression. Hum Mol Genet 2007;16:537–46. [DOI] [PubMed] [Google Scholar]
- 14. Ratnayake M, Reynard LN, Raine EV, Santibanez‐Koref M, Loughlin J. Allelic expression analysis of the osteoarthritis susceptibility locus that maps to MICAL3. BMC Med Genet 2012;13:12–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Grubbs FE. Procedures for detecting outlying observations in samples. Technometrics 1969;11:1–21. [Google Scholar]
- 16. Gee F, Clubbs CF, Raine EV, Reynard LN, Loughlin J. Allelic expression analysis of the osteoarthritis susceptibility locus that maps to chromosome 3p21 reveals cis‐acting eQTLs at GNL3 and SPCS1. BMC Med Genet 2014;15:53–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Mathelier A, Zhao X, Zhang AW, Parcy F, Worsley‐Hunt R, Arenillas DJ, et al. JASPAR 2014: an extensively expanded and updated open‐access database of transcription factor binding profiles. Nucleic Acids Res 2014;42:D142–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Messeguer X, Escudero R, Farre D, Nunez O, Martinez J, Alba MM. PROMO: detection of known transcription regulatory elements using species‐tailored searches. Bioinformatics 2002;18:333–4. [DOI] [PubMed] [Google Scholar]
- 19. Farre D, Roset R, Huerta M, Adsuara JE, Rosello L, Alba MM, et al. Identification of patterns in biological sequences at the ALGGEN server: PROMO and MALGEN. Nucleic Acids Res 2003;31:3651–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Syddall CM, Reynard LN, Young DA, Loughlin J. The identification of trans‐acting factors that regulate the expression of GDF5 via the osteoarthritis susceptibility SNP rs143383. PLoS Genet 2013;9:e1003557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bui C, Barter MJ, Scott JL, Xu Y, Galler M, Reynard LN, et al. cAMP response element‐binding (CREB) recruitment following a specific CpG demethylation leads to the elevated expression of the matrix metalloproteinase 13 in human articular chondrocytes and osteoarthritis. FASEB J 2012;26:3000–11. [DOI] [PubMed] [Google Scholar]
- 22. Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods 2012;9:671–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Rosenbloom KR, Dreszer TR, Long JC, Malladi VS, Sloan CA, Raney BJ, et al. ENCODE whole‐genome data in the UCSC Genome Browser: update 2012. Nucleic Acids Res 2012;40:D192–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hunger SP, Brown R, Cleary ML. DNA‐binding and transcriptional regulatory properties of hepatic leukemia factor (HLF) and the t(17;19) acute lymphoblastic leukemia chimera E2A‐HLF. Mol Cell Biol 1994;14:5986–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Chen H, Lin RJ, Schiltz RL, Chakravarti D, Nash A, Nagy L, et al. Nuclear receptor coactivator ACTR is a novel histone acetyltransferase and forms a multimeric activation complex with P/CAF and CBP/p300. Cell 1997;90:569–80. [DOI] [PubMed] [Google Scholar]
- 26. Takeshita A, Cardona GR, Koibuchi N, Suen CS, Chin WW. TRAM‐1, a novel 160‐kDa thyroid hormone receptor activator molecule, exhibits distinct properties from steroid receptor coactivator‐1. J Biol Chem 1997;272:27629–34. [DOI] [PubMed] [Google Scholar]
- 27. Karlsson MK, Karlsson C, Magnusson H, Coster M, von Schewelov T, Nilsson JA, et al. Individuals with primary osteoarthritis have different phenotypes depending on the affected joint: a case control study from southern Sweden including 514 participants. Open Orthop J 2014;8:450–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hermansson M, Sawaji Y, Bolton M, Alexander S, Wallace A, Begum S, et al. Proteomic analysis of articular cartilage shows increased type II collagen synthesis in osteoarthritis and expression of inhibin βA (activin A), a regulatory molecule for chondrocytes. J Biol Chem 2004;279:43514–21. [DOI] [PubMed] [Google Scholar]
- 29. Ijiri K, Zerbini LF, Peng H, Otu HH, Tsuchimochi K, Otero M, et al. Differential expression of GADD45β in normal and osteoarthritic cartilage: potential role in homeostasis of articular chondrocytes. Arthritis Rheum 2008;58:2075–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Tchetina EV, Squires G, Poole AR. Increased type II collagen degradation and very early focal cartilage degeneration is associated with upregulation of chondrocyte differentiation related genes in early human articular cartilage lesions. J Rheumatol 2005;32:876–86. [PubMed] [Google Scholar]
- 31. Wang X, Manner PA, Horner A, Shum L, Tuan RS, Nuckolls GH. Regulation of MMP‐13 expression by RUNX2 and FGF2 in osteoarthritic cartilage. Osteoarthritis Cartilage 2004;12:963–73. [DOI] [PubMed] [Google Scholar]
- 32. Mitchell PG, Magna HA, Reeves LM, Lopresti‐Morrow LL, Yocum SA, Rosner PJ, et al. Cloning, expression, and type II collagenolytic activity of matrix metalloproteinase‐13 from human osteoarthritic cartilage. J Clin Invest 1996;97:761–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Yan J, Erdem H, Li R, Cai Y, Ayala G, Ittmann M, et al. Steroid receptor coactivator‐3/AIB1 promotes cell migration and invasiveness through focal adhesion turnover and matrix metalloproteinase expression. Cancer Res 2008;68:5460–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Wang BL, Dai CL, Quan JX, Zhu ZF, Zheng F, Zhang HX, et al. Parathyroid hormone regulates osterix and Runx2 mRNA expression predominantly through protein kinase A signaling in osteoblast‐like cells. J Endocrinol Invest 2006;29:101–8. [DOI] [PubMed] [Google Scholar]
- 35. Wang CY, Yang SF, Wang Z, Tan JM, Xing SM, Chen DC, et al. PCAF acetylates Runx2 and promotes osteoblast differentiation. J Bone Miner Metab 2013;31:381–9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Scatter plots of the quantitative expression of NCOA3 (left) and SULF2 (right) in osteoarthritis (OA) cartilage. Data is stratified by (A) joint site, (B) sex and (C) age at surgery. The data points represent the average of the three replicates for each sample. The expression of each gene was assessed by quantitative real‐time reverse transcription PCR and normalized to the housekeeping genes 18S, GAPDH and HPRT1. n is the number of patients studied. The horizontal lines in the columnar scatter plots represent the mean and the standard error of the mean. The trendlines in the XY scatter plots represent linear regression of the data. p‐values for (A) and (B) were calculated using a two‐tailed Mann‐Whitney exact test. p‐values for (C) were calculated using linear regression.
Figure S2. Schematic representation of the 20q13 locus. Gene track is a screenshot from http://genome.ucsc.edu, using release hg19. The genes are represented by horizontal lines, with the arrows indicating the direction of gene transcription. The exons are represented by vertical bars, whose width is proportional to the length of the exon. The numbers above the gene tracks indicate the position within chromosome 20. The vertical lines beneath the gene tracks represent the SNPs analyzed in this study. The SNP that identified the signal in the GWAS, rs6094710, is labeled in white text on a black background, and the two transcript SNPs used for AEI, rs6094752 (NCOA3) and rs3810526 (SULF2), are labeled with bold text. Also labeled are the other SNPs in perfect LD with rs6094710.
Figure S3. Electrophoretic mobility shift assay (EMSA) analysis in SW872 (A and B) and SW1353 cells (C and D). (A and C) Increasing concentrations of unlabeled G and A allele competitor were added to the EMSA reaction containing the G and A allele probes and nuclear extract, with the arrow indicating the specific complex binding to the probes. (B and D) The addition of increasing concentrations of HLF and TCF4 unlabeled consensus competitors to the EMSA reaction containing the G or A allele probe. The arrow indicates the complex that is competed.
Figure S4. Expression of NCOA3 in cartilage tissue. (A,B,D,E) Immunohistochemical staining against NCOA3 in macroscopically normal knee (A and D) and hip (B and E) cartilage obtained from osteoarthritis patients. (C and F) Negative control staining with no primary antibody in the same hip sample as shown in B and E. The articular surface is at the top of the images. Top panels (A‐C) were taken at 5x magnification; bottom panels (D‐F) show enlarged views taken at 10x magnification. Cells exhibit nuclear staining, which is consistent with the role of NCOA3 as a co‐activator of various nuclear receptors. Enlarged regions are indicated by the black boxes in A‐C. Scale bars = 200 m.
Table S1. Table of OA patient characteristics, their genotype at rs6094710 and details of their use in quantitative real‐time reverse transcription PCR (RT‐PCR). F, female; M, male; K, knee; H, hip.
Table S2. Primer and probe sequences used for the quantitative real‐time reverse transcription PCR (RT‐PCR) of a panel of genes. The NCOA3 primers are not listed and were purchased from Applied Biosystems as an off‐the‐shelf TaqMan Gene Expression Assay. Also listed are the primers used for the pyrosequencing analysis of SNPs rs6094710, rs6094752 and rs3810526. FP, forward primer; RP, reverse primer; SP, sequencing primer; Pr, probe
Table S3. The twelve DNA fragments examined by luciferase analysis. Chromosome locations based on UCSC genome browser, release hg19. The sites in the cloning primers for the restriction enzymes MluI (ACGCGT) and XhoI (CTCGAG) are highlighted in bold.
Table S4. Electrophoretic mobility shift assay (EMSA) probes and competitors. The bold and underlined bases in the EMSA probes indicate the position of rs116855380. The series of underlined bases in the transcription factor competitors indicate the site of the consensus sequence for each factor.
Table S5. Antibodies used for supershift electrophoretic mobility shift assays


