Figure 2.
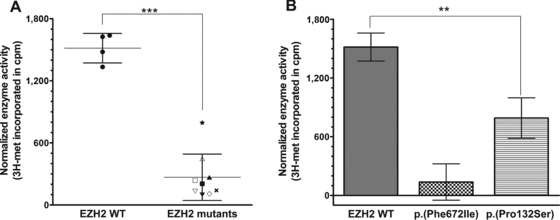
Weaver syndrome mutants are impaired in their histone methyltransferase activity in vitro. Histone methyltransferase reactions were performed using 2 μg purified core histones and 0.67 μM 3H‐S‐adenosyl‐methionine (3H‐SAM). Each reaction was incubated with 250 ng of either wild‐type (WT) or a mutant HMTase complex (or no enzyme controls). Histone methyltransferase activity was measured based on the incorporation of 3H‐labeled methyl groups, represented in scintillation counts per minute. Counts were normalized by subtracting background counts (i.e., no enzyme) from the total counts. A: Incorporation of tritiated methyl groups from 3H‐SAM onto core histones is shown for each complex: EZH2 WT •, p.(Phe672Ile) ×, p.(Pro132Ser) ★, p.(Tyr153del) △, p.(His694Tyr) ▽, p.(Glu745Lys) ▴, p.(Ala682Thr) ▾, p.(Arg684Cys) ▪, p.(Tyr133Cys) □, and p.(Asp185His) ◇. Error bars represent standard deviation (SD) within the groups “EZH2 WT” and “EZH2 mutants.” Unpaired t‐test showed statistically significant difference between the two groups (P value < 0.0001). B: Incorporation of tritiated methyl groups from 3H‐SAM onto core histones is shown for the positive control EZH2 WT, the negative control EZH2 (p.Phe672Ile), and the mutant complex with activity closest to WT, namely, EZH2 (p.Pro132Ser). Error bars represent SD of four independent replicates for the controls, and three independent replicates for the mutant EZH2 (p.Pro132Ser). One‐way ANOVA showed statistically significant difference between all groups (overall P value < 0.0001; P values between WT and p.(Phe672Ile), between p.(Phe672Ile) and p.(Pro132Ser), and between WT and p.(Pro132Ser) were all <0.05).
