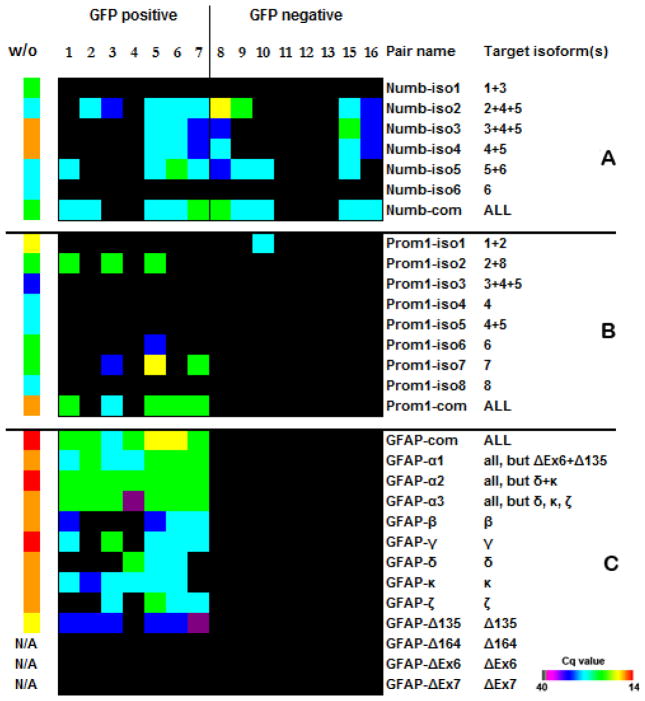
Figure 4. Expression patterns of Numb, Prominin1, and GFAP isoforms.
(A) Numb isoforms 1, 3, and 6 that were not detected contain insertions into the PRR domain, and the detected Numb isoforms 2, 4, and 5 do not have an insertion (36). Because the same pair of primers can detect different isoforms, it is not possible to assign the expression of a certain isoform to a specific cell. For example, cells 5–8 can express either the combination of isoforms 2, 4, and 5 or only isoform 5. At the same time, Numb2 is expressed in cells 2 and 3, Numb4 in cell 16, and Numb5 in all cells where it was detected. (B) The cells expressed mostly Prominin1 isoforms 2 and 7. No isoform-specific transcripts were detected in cell #6, but primers recognizing all isoforms produced a signal. Our results also show good concordance with previous studies that revealed the expression of only Prominin1 transcript isoforms 1, 2, and 6 in the subventricular zone (SVZ), and the absence of isoforms 3, 4, 5, and 8 (34,37). We also observed that isoform 1 was expressed exclusively in one of the GFP− cells, while other isoforms were present only in GFP+ population. While the expression of isoform 7 was not previously assessed, we found that this isoform together with isoform 2 is dominant in GFP+ cells. Prominin1 is strongly predisposed to alternative splicing, and there are 10 known isoforms for the human transcript. Therefore, it is likely to reveal a new murine isoform of Prominin1 amidst the eight known isoforms. (C) Glial fibrillary protein (GFAP) expression was previously detected in distinct population of astrocytes that have been identified as SVZ neural stem cells (38–40). GFAP isoforms were detected only in GFP+ cells. The cells analyzed in our experiments did not contain isoforms GFAP-ΔEx6, GFAP-ΔEx7, and GFAP-Δ164, and the GFAP-Δ135 isoform was expressed at a low level. Isoforms GFAP-ΔEx6, GFAP-Δ164 and GFAP-Δ135 were not detected in samples without amplification (w/o), but the quality of the primers was verified before (39). Primers labeled “com” are designed to anneal to the common part of all known isoforms of the transcript. Because some isoforms are homologous, certain primers detect more than one target isoform. Heat maps reflect real-time PCR-derived Cq values for each transcript. A sample without amplification was used as positive control. Total RNA input of unamplified, pooled SVZ cells or embryonic cells in the RT reaction was 3 μg.