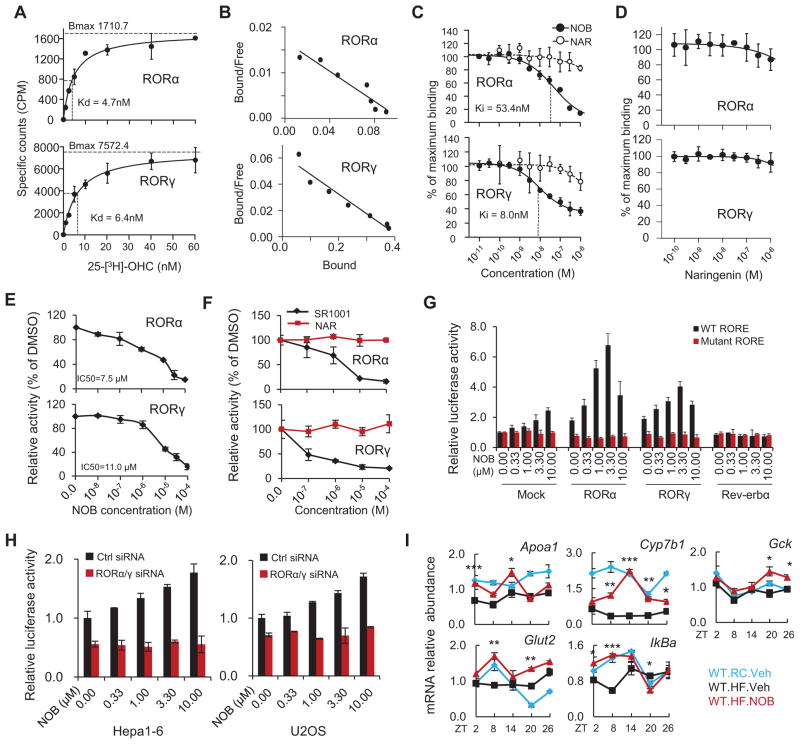
Figure 6. NOB enhanced RORα/γ transcriptional activity via direct binding to RORα/γ.
(A and B) Saturation curves and Scatchard plots from filter binding assays for RORα-LBD and RORγ-LBD. (A) Saturation curves for 25-[3H]-OHC binding were generated with 100ng of RORα-LBD (top) and 200ng of RORγ-LBD (bottom) (n=3). Dissociation constant values are shown. (B) Saturation curve results in Fig. 5E were subjected to Scatchard analysis and Scatchard plots for 25-[3H]-OHC are shown for RORα-LBD (top) and RORγ-LBD (bottom) corresponding to (n=3). This analysis gave a dissociation constant (Kd) of 6.10 nM and a total number of binding sites (Bmax) of 100 fmol/mg of protein for RORα, and 6.67 nM and 410 fmol/mg of protein for RORγ. (C and D) In vitro competitive radio-ligand binding assay indicating the direct binding of NOB (C), but not NAR (C) or Naringenin (D), to RORα-LBD and RORγ-LBD within the indicated dose range. Inhibitory constant values are shown. (E and F) Mammalian one-hybrid assays showing ligand interaction with ROR-LBD. HEK293T cells were cotransfected with a GAL4 reporter construct with expression vectors for GAL4 DBD-RORα LBD or GAL4 DBD-RORγ LBD. Cells were treated with varying concentrations of NOB (E) and its non-methoxylated analog NAR (F). SR1001 served as a positive control in (F). (G) NOB dose-dependently increased Bmal1 promoter-driven luciferase reporter expression with wild-type, but not mutant, RORE in the presence of RORα or RORγ in Hepa1-6 cells. (H) Knockdown of RORα/γ expression by siRNAs abrogated NOB induction of Bmal1 promoter-driven luciferase reporter expression in both Hepa1-6 and U2OS cells. (I) Real-time qPCR analysis of RORα/γ target genes from the same mouse liver samples as in Figure 5A. Data are presented as mean ± SEM. *p < 0.05, **p < 0.01, and ***p < 0.001. WT.HF.NOB vs. WT.HF.Veh.