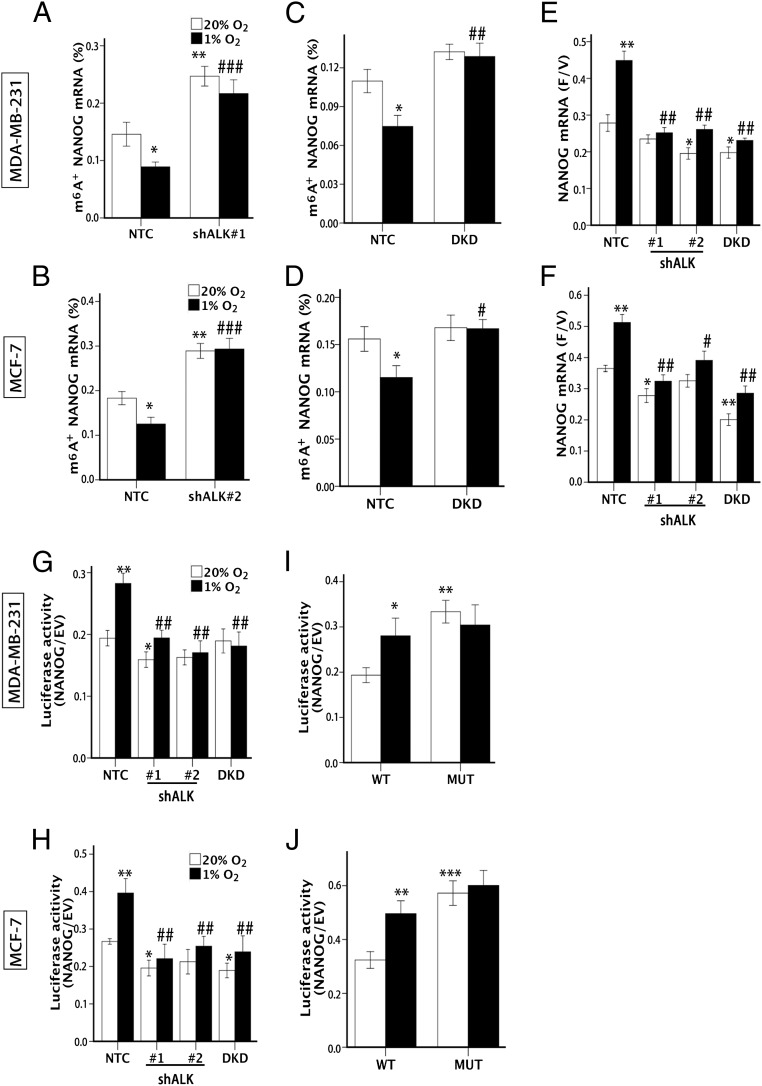
Fig. 3.
ALKBH5 demethylates and stabilizes NANOG mRNA. (A and B) MDA-MB-231 (A) and MCF-7 (B) NTC and shALK subclones were exposed to 20% or 1% O2 for 48 h. m6A immunoprecipitation and RT-qPCR were performed to determine the percentage of NANOG mRNA with methylation (m6A+; mean ± SEM; n = 3; *P < 0.05 and **P < 0.01 vs. NTC at 20% O2; ###P < 0.001 vs. NTC at 1% O2). (C and D) MDA-MB-231 (C) and MCF-7 (D) NTC and DKD subclones were exposed to 20% or 1% O2 for 48 h, and the percentage of m6A+ NANOG mRNA was determined (mean ± SEM; n = 3; *P < 0.05 vs. NTC at 20% O2; #P < 0.05 and ##P < 0.01 vs. NTC at 1% O2). (E and F) MDA-MB-231 (E) and MCF-7 (F) subclones were exposed to 20% or 1% O2 for 24 h and treated with vehicle or flavopiridol for 3 h (E) or 6 h (F), and NANOG mRNA levels were measured by RT-qPCR and the F/V ratio of NANOG mRNA was determined (mean ± SEM; n = 3; *P < 0.05 and **P < 0.01 vs. NTC at 20% O2; #P < 0.05 and ##P < 0.01 vs. NTC at 1% O2). (G and H) Luciferase assays were performed by transfecting MDA-MB-231 (G) and MCF-7 (H) subclones with a reporter plasmid containing luciferase coding sequences followed by the NANOG-3′UTR or EV containing luciferase only. The ratio of luciferase activity in cells transfected with NANOG-3′UTR relative to EV was determined (mean ± SEM; n = 3; *P < 0.05 and **P < 0.01, vs. NTC at 20% O2; ##P < 0.01 vs. NTC at 1% O2). (I and J) Luciferase assays were performed in MDA-MB-231 (I) or MCF-7 (J) cells transfected with WT or mutant (MUT) luciferase-NANOG-3′UTR reporter (mean ± SEM; n = 3; *P < 0.05, **P < 0.01, and ***P < 0.001 vs. NTC at 20% O2).