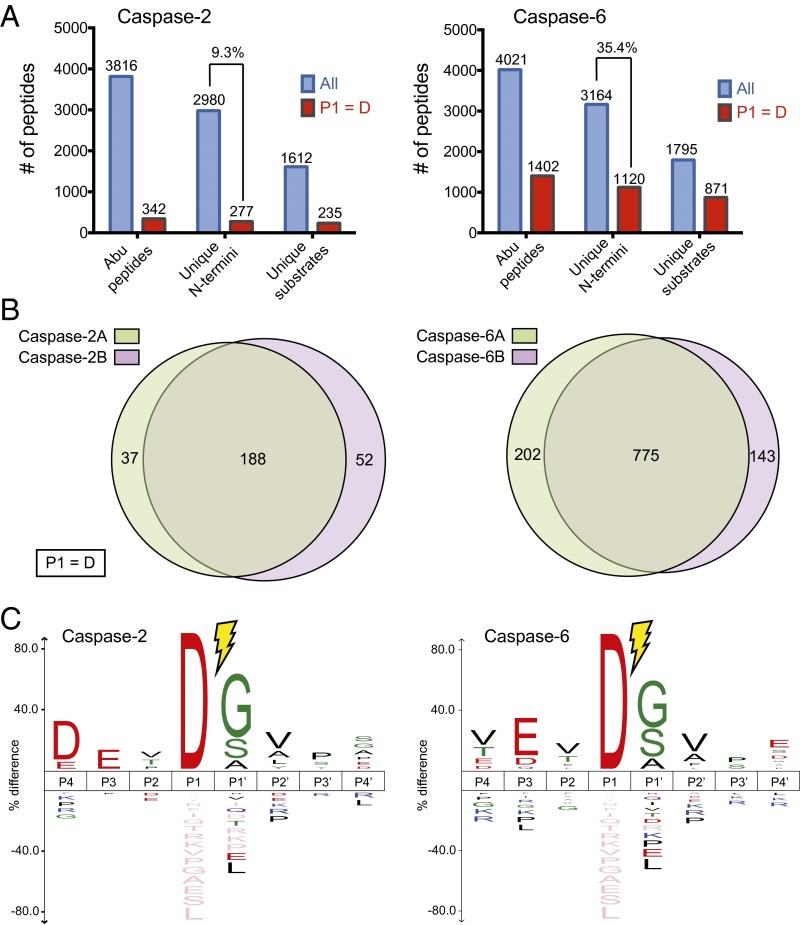
Fig. 2.
Discovery of caspase-2 and caspase-6 substrates in cell lysates. (A) Shown are the total number of unique peptides (tagged with Abu), unique cleavage sites, and unique substrates at the protein level identified for caspase-2 (Left) and caspase-6 (Right). The subset of identifications featuring an Asp at P1 position is shown in red. Total peptides are colored in blue, whereas peptides originating only from a caspase cleavage event (aspartate at P1 position) are colored in red. (B) The discovery experiments show greater than 70% overlap between the two biological replicates in the cleavage sites identified for both caspase-2 and caspase-6. (C) Sequence recognition motifs of caspase cleavages for caspase-2 and caspase-6 shows highest consensus DEVD↓(G/S/A) and VEVD↓(G/S/A) motifs, respectively (P value = 0.05). Consensus sequences were generated using iceLogo (24).