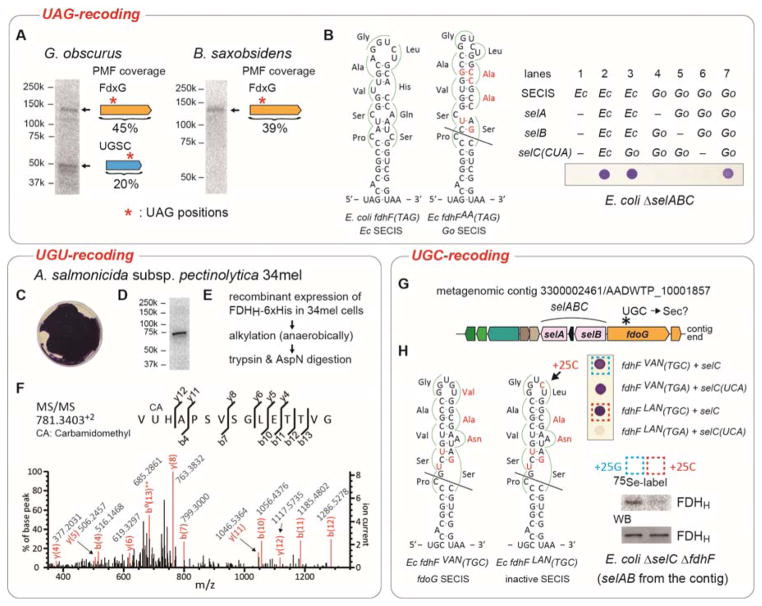
Figure 2. Recoding of UAG and cysteine codons to selenocysteine.
(A) Metabolic 75Se labeling of G. obscurus and B. saxobsidens cells. Crude extracts were resolved by SDS-PAGE, and their putative selenoproteins were visualized by PhosphorImager analysis. The results of peptide mass fingerprinting (PMF) analyses of the proteins in the excised gel bands are shown to the right of the bands. (B) FDHH expression in E. coli ΔselABC ΔfdhF cells with the G. obscurus selABC genes and a chimeric fdhF(140TAG) gene variant having a G. obscurus SECIS element with a few nucleotide modifications shown in red. The selC(CUA) genes express tRNASecCUA. The expressed selenoprotein FDHH reduced benzyl viologen, resulting in a purple color. (C) FDHH activity of A. salmonicida subsp. pectinolytica 34mel cells. (D) Metabolic 75Se labeling of the FDHH of 34mel. (E) The procedure of sample preparation for the LC-MS/MS analysis of the FDHH selenoprotein. (F) PMF confirms Sec incorporation at codon 140 in the recombinant FDHH. (G) tRNASecGCA gene (selC) locus in a metagenomic contig. (H) In vivo FDHH assays in E. coli ΔselABC ΔfdhF cells with the selABC genes of the metagenomic contig and a chimeric fdhF(140TGC) gene carrying the contig’s SECIS element with a few nucleotide modifications shown in red. The two transformed strains boxed were metabolically labeled with 75Se, and the radioactive FDHH proteins were analyzed by SDS-PAGE and autoradiography or western blotting (WB).