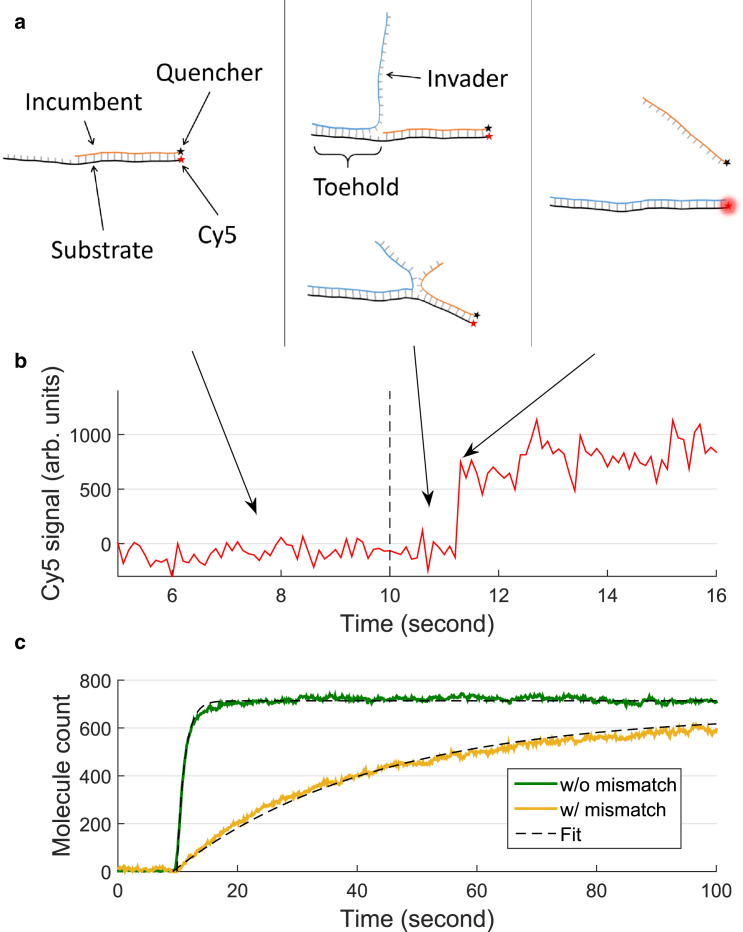
Figure 1.
Measuring strand displacement. (a) Experimental design. Three single strands of DNA termed substrate (black), incumbent (orange), and invader (blue) strands participate in strand displacement. Cy5 attached to the substrate is initially quenched due to the black hole quencher on the incumbent. When the incumbent is displaced by the invader, Cy5 recovers its fluorescence. (b) Cy5 signal during strand displacement. Shown is the fluorescence time trajectory of a single Cy5 molecule obtained by total internal reflection microscopy. Invader molecules were introduced via flow (dashed line at 10 s). A large, single, and sudden increase in fluorescence indicates displacement. (c) Extracting the apparent strand displacement rate. Two sets of sample data and their respective fits are plotted. Molecule count is calculated via in-house code scripted in MATLAB (The MathWorks). The data are fitted to single exponential curves with an origin at the injection time (10 s after starting acquisition). MFPTs are approximated as the reciprocal of the fitted rate constants. To see this figure in color, go online.