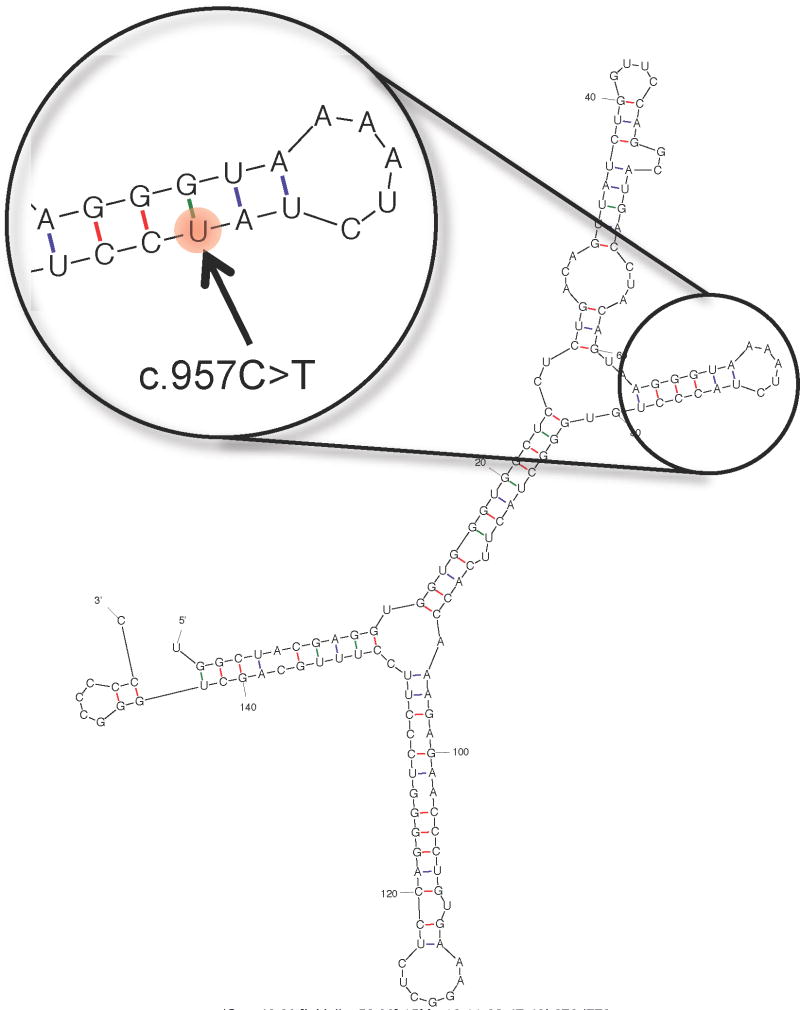
Figure 2.
Partial view of the secondary structure of the CDSN transcript. The structure was predicted using mfold (http://mfold.rna.albany.edu/?q=mfold) and 151 bps (corresponding to coordinates chr6:31084360-31084510 in the hg19 assembly of the human genome) centered on the position of the synonymous mutation c.957C>T (rs1062470). The structure corresponding to the synonymous mutation is predicted to be identical to the wild-type structure (the mutated position is highlighted in the enlarged inset), with the exception of the level of free energy, which is slightly lower for the structure corresponding to the reference C allele (ΔG = −46.80 kcal/mol) relative to the structure corresponding to the synonymous T variant (ΔG = −44.20 kcal/mol).