Figure 2.
Live Imaging of Zscan4 and Rex1
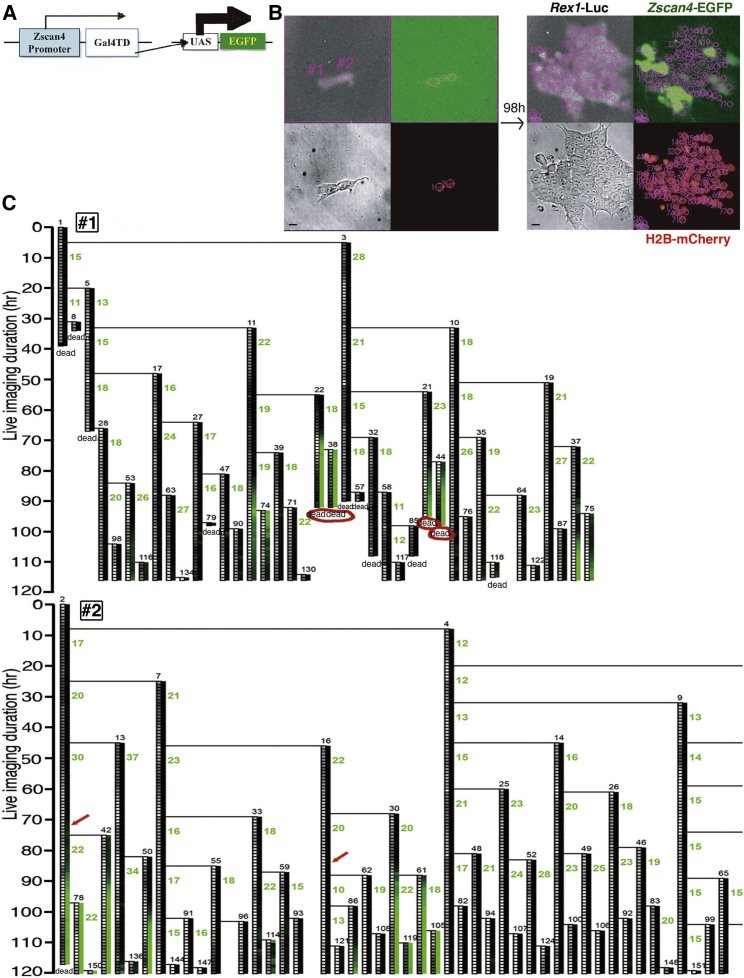
(A) Design of the Zscan4 reporter transgene. Upon the activity of the Zscan4 promoter, the trans-activator domain of GAL4 is expressed, which trans-activates the co-transfected UAS promoter with EGFP downstream. See also Figure S4A.
(B) Example of cell tracking starting from two cells (#1 and #2). mCherry fused to H2B was used as a nuclear marker. Images were captured every 60 min up to 120 hr. A region of the nucleus in each cell was drawn manually and intensities of Luciferase2 (for Rex1) and GFP (for Zscan4) were measured. Bars, 10 μm.
(C) Lineage trees generated from the cell tracking of cells #1 and #2 in (B). Green numbers indicate the cell-cycle length (hr). Intensities of Luciferase and GFP (described in Figures S2E and 3A, respectively) are shown by color scales. Input data and the program to draw the lineage trees are provided in Data S1. Red arrows indicate subtle GFP intensities, and red circles indicate cell death in GFP-strong-positive cells (both are discussed in Figure 3B). During culture, cells seemed to die randomly. However, from the lineage tree, we noticed that most of the dead cells derived from cell #1. This means that the death fate was already determined in the two apparently identical cells shown in (B) (#1 and #2). See also Figures S1–S3for Rex1.