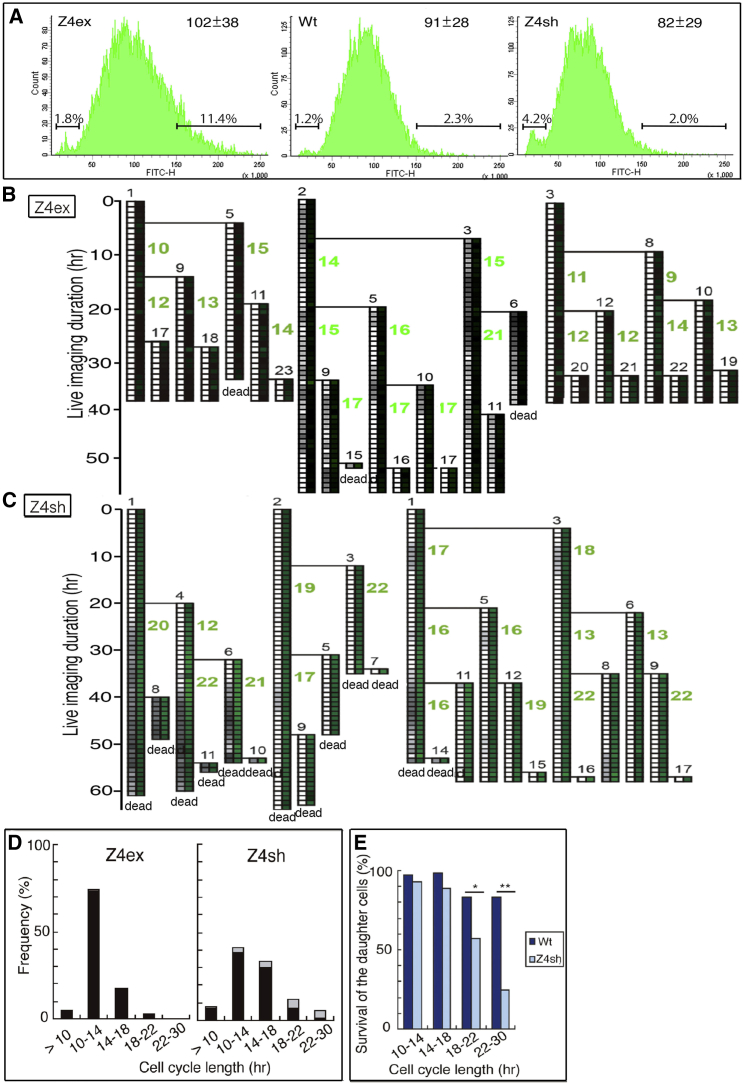
Figure 6.
Expression Levels of Zscan4 Affects the Cell-Cycle Length
(A) Telomere lengths of Z4ex-, wild-type or Z4sh-ESCs were measured by flow-FISH using an FITC-conjugated telomere probe. The average telomere intensity (×1,000) per cell ±SD is indicated. n > 7,000 cells. Two technical replicates showed the same tendency. Note that cells with longer telomeres were increased in Z4ex-ESCs compared with wild-type ESCs (11.4% vs 2.3%) and cells with shorter telomeres were increased in Z4sh-ESCs compared with wild-type ESCs (4.2% vs 1.2%). See also Figures S6B and S6C.
(B and C) Examples of the lineage trees of Z4ex-ESCs (B) and Z4sh-ESCs (C) with Rex1-Luciferase and Zscan4-Gal4-UAS-EGFP probes. The green scale on the right side of each lineage indicates the intensities of the Zscan4-Gal4-UAS-EGFP. Note that Z4ex-ESCs showed stable cell cycles without strong Zscan4 activities (B), while Z4sh-ESCs showed a longer cell cycle with high basal EGFP expression (C).
(D) Histograms of the cell-cycle length of Z4ex- and Z4sh-ESCs (n = 194 and 289 cell divisions in three and 11 lineages from two independent experiments, respectively). Z4ex-ESCs increased in cells with shorter cell cycles. Black, the cells whose daughter cells divided within 30 hr; gray, the cells whose daughter cells died or did not divide within 30 hr.
(E) Equivalent of the histograms of the wild-type and Z4sh-ESCs from Figures 1B and (D), respectively, highlighting the difference in the survival rate of the daughter cells, i.e., black portion divided by black + gray. Student’s t test was used for statistical analysis. ∗p < 0.01, ∗∗p < 0.005. See also Figure S7.