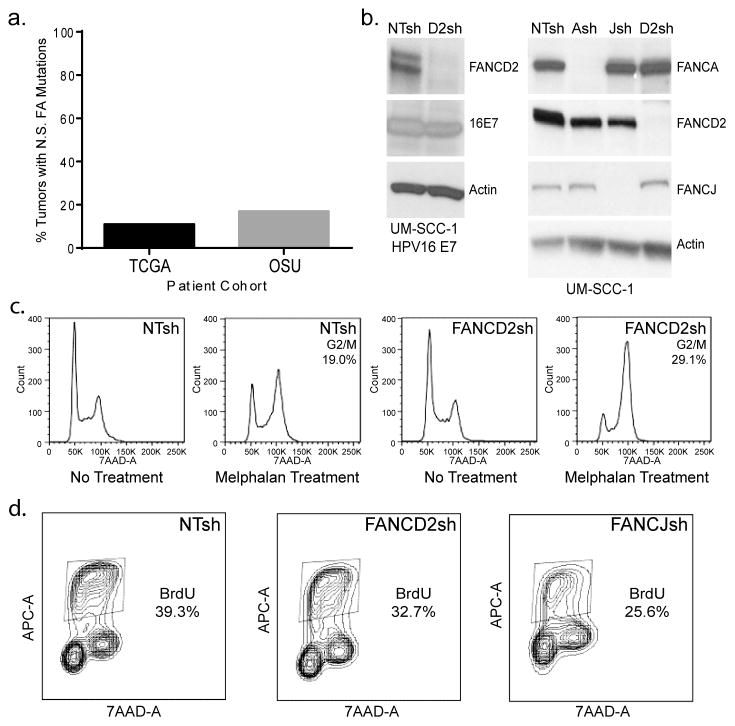
Figure 1. Generation of FA deficient, HPV positive and negative, HNSCC cell models.
(a) The somatic mutations table (MAF file) for head and neck squamous carcinoma (HNSC) samples was obtained from TCGA data portal and from an independent cohort of sporadic HNSCC tumors from Ohio State University (OSU). Analysis of mutational data from 306 (TCGA) and 34 (OSU) sporadic HNSCC tumors determined that 11.1% and 17.6%, respectively, harbored non-synonymous (N.S.) mutations in one of 16 FA genes, respectively. (b) UM-SCC1 HNSCC (HPV16 E7-positive and –negative) cells were knocked down for FANCA, FANCD2 and FANCJ, followed by western blot analysis for verification of protein depletion. (c) FA knockdown by shRNA transduction leads to classical FA phenotypes. UM-SCC1 cells were treated with melphalan, and subjected to flow cytometry based cell cycle analysis. The numbers listed indicate percentages of cells in G2/M following melphalan exposure. FANCD2-deficient cells were increased for the proportion of cells in G2/M when compared to the NTsh control cell population. (d) BrdU incorporation in FANCD2- and FANCJ-deficient compared to control UM-SCC1 cells reveals a slight decrease in proliferation.