Figure 4.
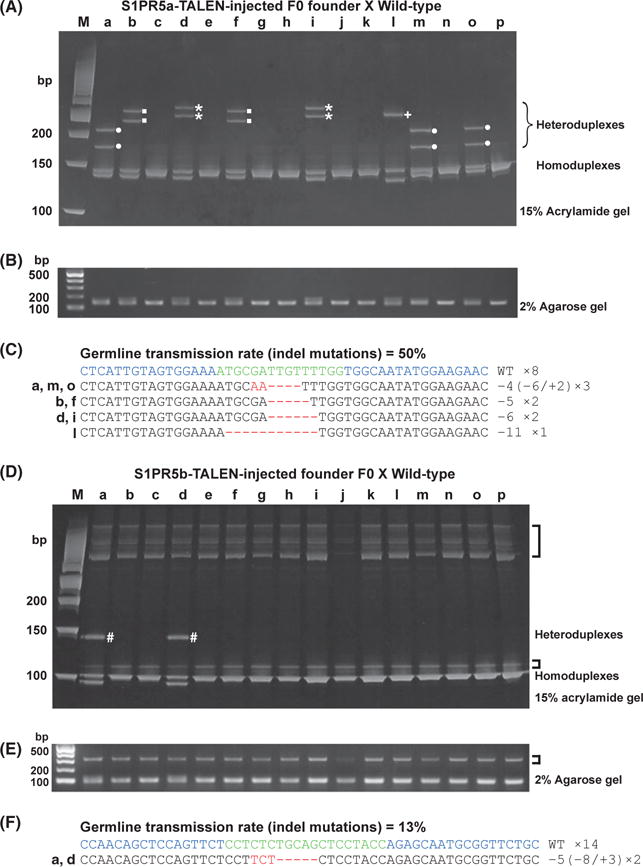
Detection of S1PR5a and S1PR5b mutant alleles by HMA in F1 embryos. (A–C) S1PR5a mutant alleles. (D–F) S1PR5b mutant alleles. (A, D) PCR amplicons from individual F1 genomic DNAs were electrophoresed on a 15% polyacrylamide gel. For S1PR5a, four distinct HMA profiles were detected (●, ■, ⋆, +); for S1PR5b, only one unique HMA profile was detected (#). (B, E) Band patterns of PCR fragments from S1PR5a (B) and S1PR5b (E) target regions were similar in all lanes of a 2% agarose gel. Nonspecific bands amplified by S1PR5b primer sets were observed in all samples (brackets). (C, F) PCR amplicons were subcloned into a pGEM-T Easy vector, and inserts from individual F1 embryos were sequenced. Deleted and inserted nucleotides in the DNA sequences are indicated by red dashes and red letters, respectively. The wild-type sequence is shown at the top. The number of nucleotides deleted (−) and inserted (+) is indicated to the right with the detection number. The germ-line transmission rate is indicated at the top. Blue: TALEN target sequences. Green: spacer sequences. (HMA, heteroduplex mobility assay; TALEN, transcription activator-like effector nucleases).
