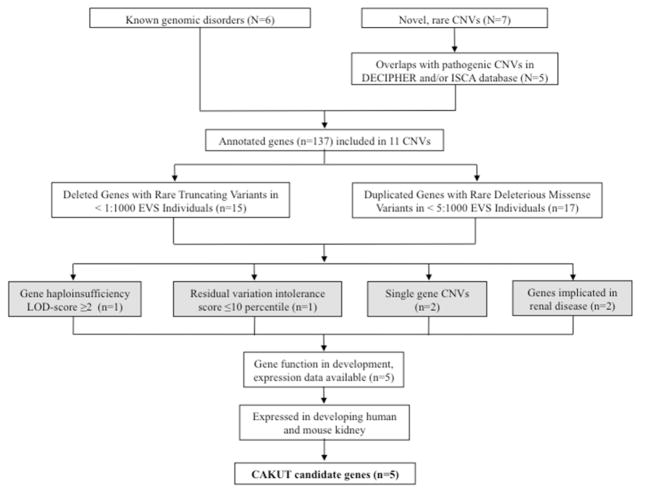
Figure 1. From CNVs to candidate genes for CAKUT.
All identified CNVs (N) were included in the analysis. For all novel, rare CNVs, deletions and duplications that showed significant overlap to pathogenic or uncertain pathogenic CNVs in public databases were included (see Supplementary Figure 1 and Supplementary Table 6). After annotation of gene content (n), genes that displayed rare truncating variants (deletions) and rare missense variants (duplications) in the Exome Variant Sever Database (http://evs.gs.washington.edu/EVS) were selected. We then assessed haploinsufficiency (HI-)LOD-scores and residual variation intolerance (RVI) scores for the prioritized genes (threshold values: HI LOD ≥2 and/or RVI-score <10th percentile) and included the prioritized genes within single gene CNVs as well as those genes that are implicated in renal disease. One gene met >1 threshold value for inclusion (DLG1). Gene expression profiles in the developing mouse kidney for all high-priority genes were evaluated by using GUDMAP (http://www.gudmap.org) and Genepaint (http://www.genepaint.org/) databases. Finally, we performed immunofluorescence studies in an E14.5 mouse kidney. By using this systematic bioinformatic approach, we prioritized 5 candidate genes for CAKUT.
CAKUT, congenital anomalies of the kidney and urinary tract; CNV, copy number variation and LOD, logarithm of the odds. Web-resources: Database of Chromosomal Imbalance and Phenotype in Humans Using Ensembl Resources Consortium (DECIPHER; http://decipher.sanger.ac.uk/); International Standards For Cytogenomic Arrays Consortium (ISCA; https://www.iscaconsortium.org/).