Figure 1. The biosynthesis of Moco.
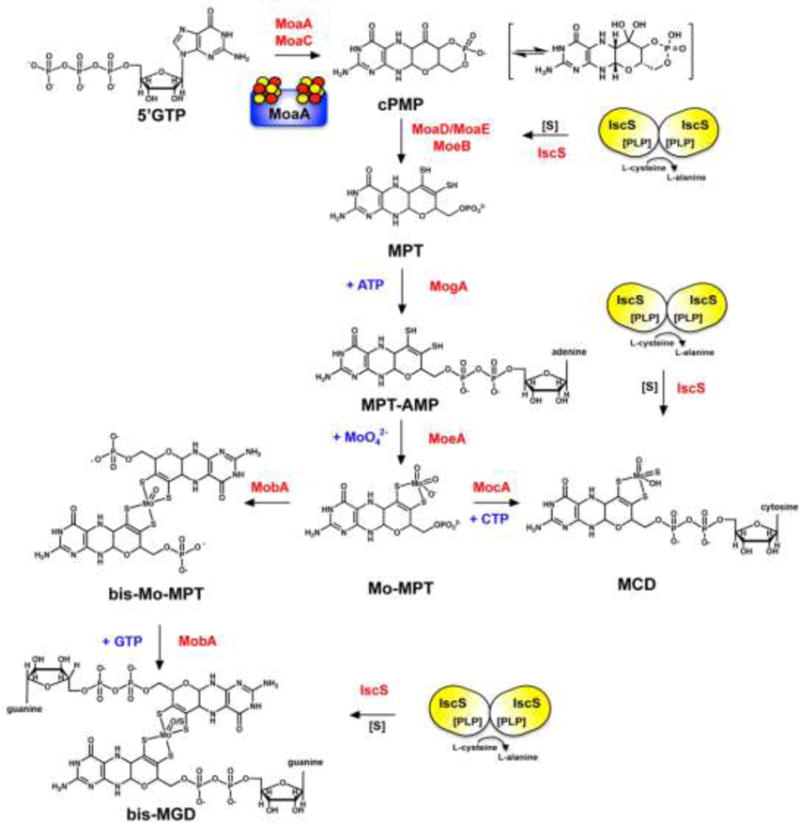
Shown is a scheme of the biosynthetic pathway for Moco biosynthesis in bacteria and the proteins involved in this pathway (which were mainly identified by studies using the E. coli proteins). Mo-MPT is formed from 5′GTP with cPMP, MPT and MPT-AMP as intermediates. The first reaction involves an FeS cluster containing protein (schematically the MoaA protein is shown containing two 4Fe4S clusters). Mo-MPT can be further modified by formation of an bis-Mo-MPT intermediate, and further addition of a GMP molecule to each MPT unit, forming the bis-MGD cofactor. Both reactions are catalyzed by the MobA protein. Bis-MGD can be further modified by the addition of a sulfido-ligand at the Mo-active site, a reaction catalyzed by the L-cysteine desulfurase IscS. Alternatively, Mo-MPT is modified by the addition of CMP to form the MCD form of the cofactor. Here, a terminal sulfur ligand is added to the molybdenum site, generating sulfurated MCD, a reaction catalyzed by the L-cysteine desulfurase IscS. The names of the proteins involved in the reactions are colored in red, nucleotides required and molybdate are colored in blue.
