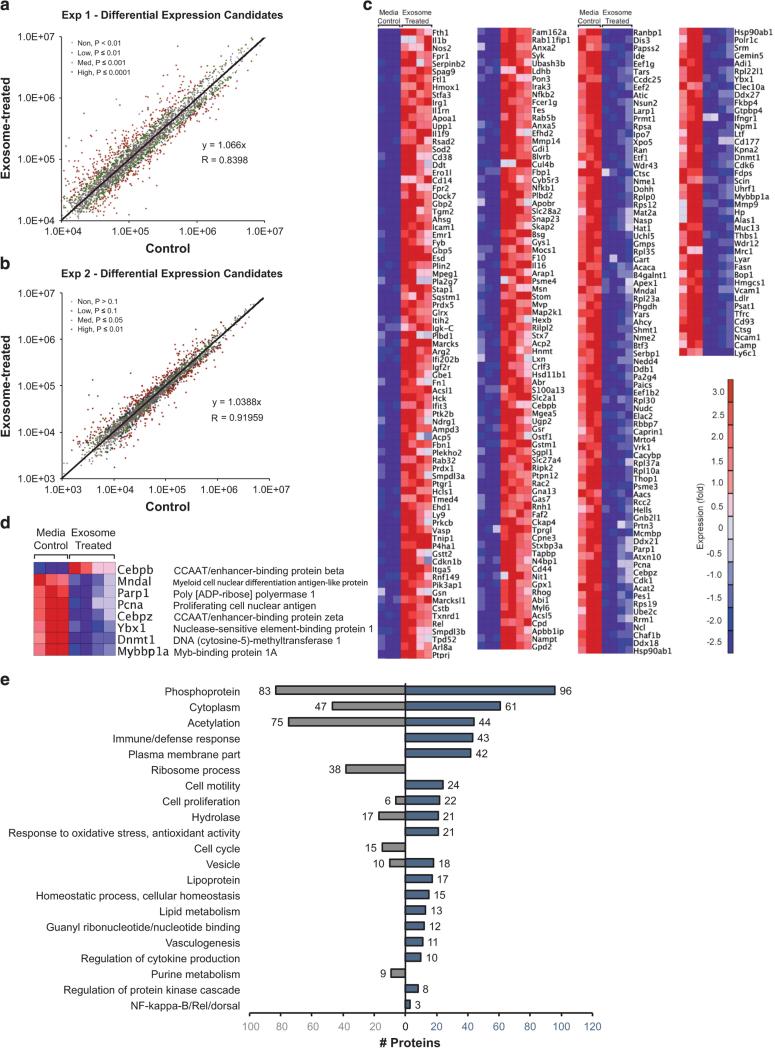
Figure 5.
Differentially expressed proteins and gene ontology. We performed two TMT experiments to compare proteins in exosome-treated HSPC, compared with controls. From five control and eight exosome-treated samples across 2 experiments, we obtained 282 overlapping proteins differentially expressed (P ≤ 0.0001 for Exp. 1, P ≤ 0.10 for Exp. 2) between exosome- treated and control samples. Because of higher sampling of peptides, Exp. 1 yielded the highest quality data and was thus subjected to more stringent statistical criteria. (a and b) Averaged summed reporter ion intensities for proteins in exosome-treated versus control samples used for a normalization check of data from Exp. 1 and Exp. 2, respectively, showing differentially abundant proteins in color with indicated P-values. (c) Heat map of differentially expressed proteins in Molm-14 exosome-treated versus control HSPC from Exp. 1. (d) Relative protein expression of differentially expressed transcription factors. (e) Up- and downregulated gene ontology processes from DAVID (P < 0.001; see Supplementary File 1) for differently expressed proteins in exosome-treated versus control HSPC.