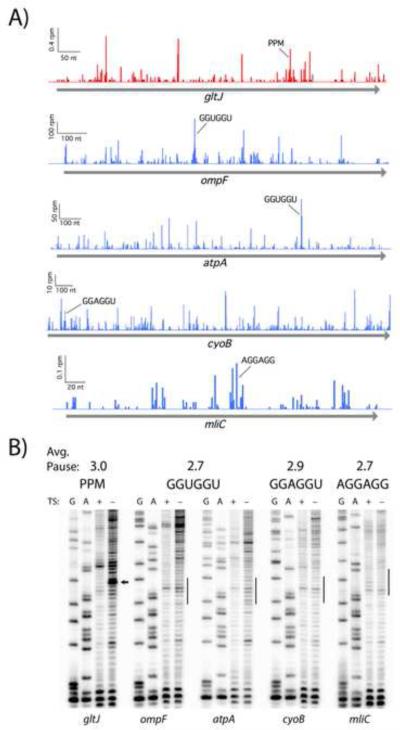
Figure 3.
Pauses at Shine-Dalgarno motifs are not detected in an in vitro translation assay. A) SD pauses appear in profiling data from MG1655 (Li et al. 2012, blue). Likewise, pauses appear at Pro-Pro-Met (PPM) in a mutant lacking EFP (Woolstenhulme et al. 2015, red). B) Toeprinting analysis of four strong SD motifs and a Pro-Pro-Met control with roughly equivalent pause scores in ribosome profiling data. Expected pausing sites are indicated with an arrow or line. Thiostrepton (TS) traps the ribosome at start codons: bands seen in both treated and untreated lanes are reverse transcriptase artifacts whereas true toeprints appear in only the untreated lane.