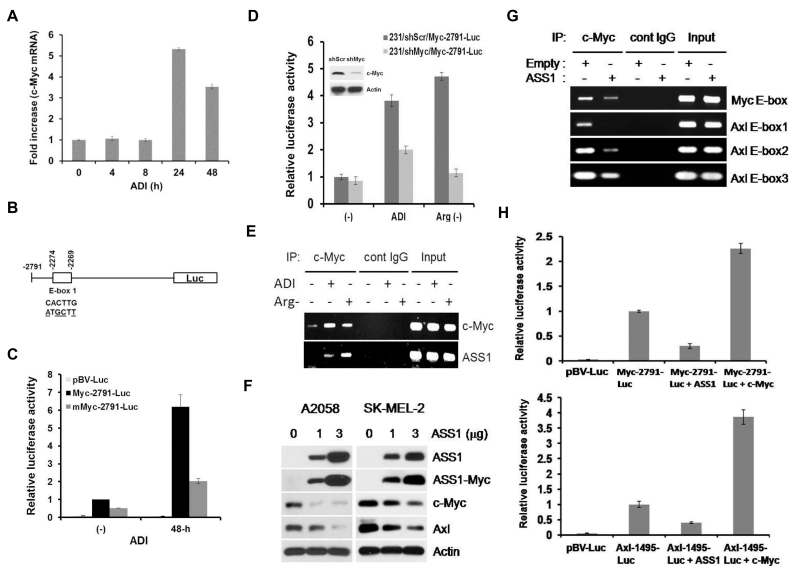
Figure 6.
Inter-regulatory network in Arg-auxotrophic response. A, quantitative real-time PCR measurement of c-Myc mRNA in A2058 cells treated with ADI for the time intervals as indicated. Results are from three independent experiments. B, schematic of the c-Myc-2791-luc construct in which wild-type and mutant E-boxes sequences are indicated. C, E-box-dependent activation of c-Myc promoter by ADI in A2058 cells. D, effects of c-Myc shRNA on the c-Myc promoter activity by ADI in the reporter assay in MDA-MB-231 cells. Cells were transfected with c-Myc shRNA or scramble shRNA (shScr) for 24 hr and treated with ADI or grown in Arg-free medium for 24 hr. Luc activities were determined from cell lysates. E, ChIP assay of c-Myc binding to the c-Myc promoter in 2058 cells treated with Arg-deprivation conditions. Input, genomic DNA prior to IP. F, inhibition of c-Myc and Axl expression by overexpressed ASS1 in A2058 and SK-MEL-2 cells. Cells were transfected with 1 and 3 μg of ASS1-Myc-Tag recombinant for 24-hr. Expression levels of ASS1, ASS1-Myc-tag, c-Myc, Axl, and Actin were analyzed by Western blotting. G, suppression of c-Myc binding to the E-box and 3 E-boxes located at the c-Myc and the Axl promoter in A2058 cells as determined by ChIP assay. H, inhibition of c-Myc (upper) and Axl (lower) promoter activity by overexpressed ASS1 in transfection assay. The ASS1-expressing recombinant was co-transfected with Myc-2791-Luc, Axl-1494-luc, or empty vector pGV-luc into MDA-MB-231 cells. Expression of the reporters was measured and normalized to those without overexpression ASS1 as 1. Error bars represent standard deviations from three independent experiments. Melanoma cells A2058 or SK-MEL-2 cells were used here because of their low basal ASS1 expression.