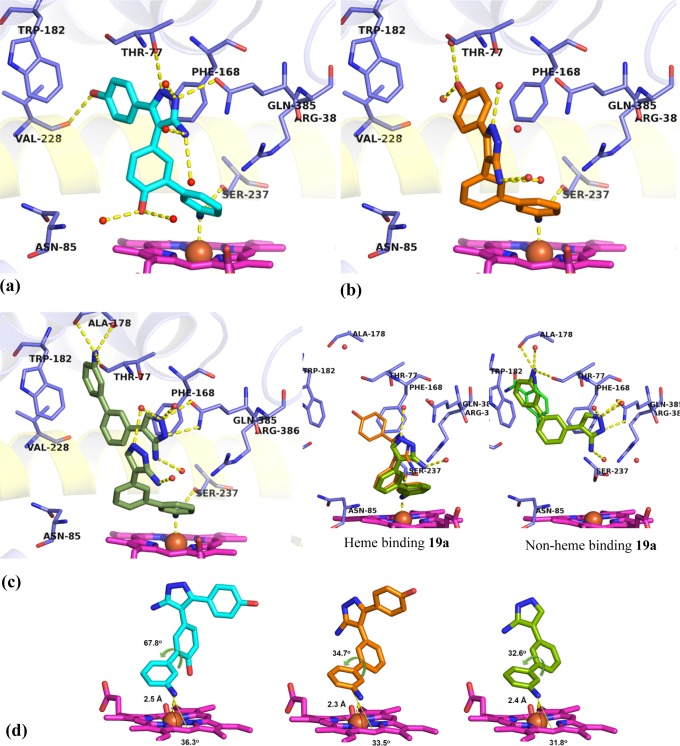
Figure 7.
X-ray crystal structures of heme binding Ar2 analogues 25b (PDB 5IBJ), 25a (PDB 5IBE), and 19a (PDB 5IBF) in complex with CYP121. (a) 25b (cyan) closely recapitulated the binding of lead 2, forming polar contacts (yellow dashes) with amino acids (blue sticks) in binding hotspots as well as additional binding contacts to Ser237, Val228, and the heme iron. (b) 25a (orange) was rotated away from the I-helix (yellow cartoon), placing the 5-aminopyrazole ring in the Ar3 pocket. Polar contacts to the heme iron and Ser237 are indicated (yellow dashes). (c) Two molecules of 19a bound per active site, each ligand fulfilling a subset of the interactions identified as binding hotspots. Inset: Heme binding 19a recapitulated the binding mode of 25a while the non-heme binding 19a ligand satisfied aromatic interactions in the Ar1 pocket, as well as the hydrogen bonding interactions of fragment 7. (d) Extracted structures of 25b, 25a, and 19a annotated to show metal coordination (yellow dashes) distance, angle of approach to the porphyrin ring (yellow angle), and dihedral torsion in the biphenyl-aniline motif. Removal of the Ar2 4-hydroxy group reduced the dihedral angle of the biphenyl aniline system in 19a and 25a by 30° compared to 25b and allowed the analogues to approach the plane of the porphyrin at a sharper angle (31–33°). The omit Fo – Fc electron density maps of ligands 19a, 25a, and 25b contoured to 3σ have been provided in Supporting Information Figure S5. Figures prepared using PyMOL v1.7.4 (Schrödinger, LLC).