Figure 1.
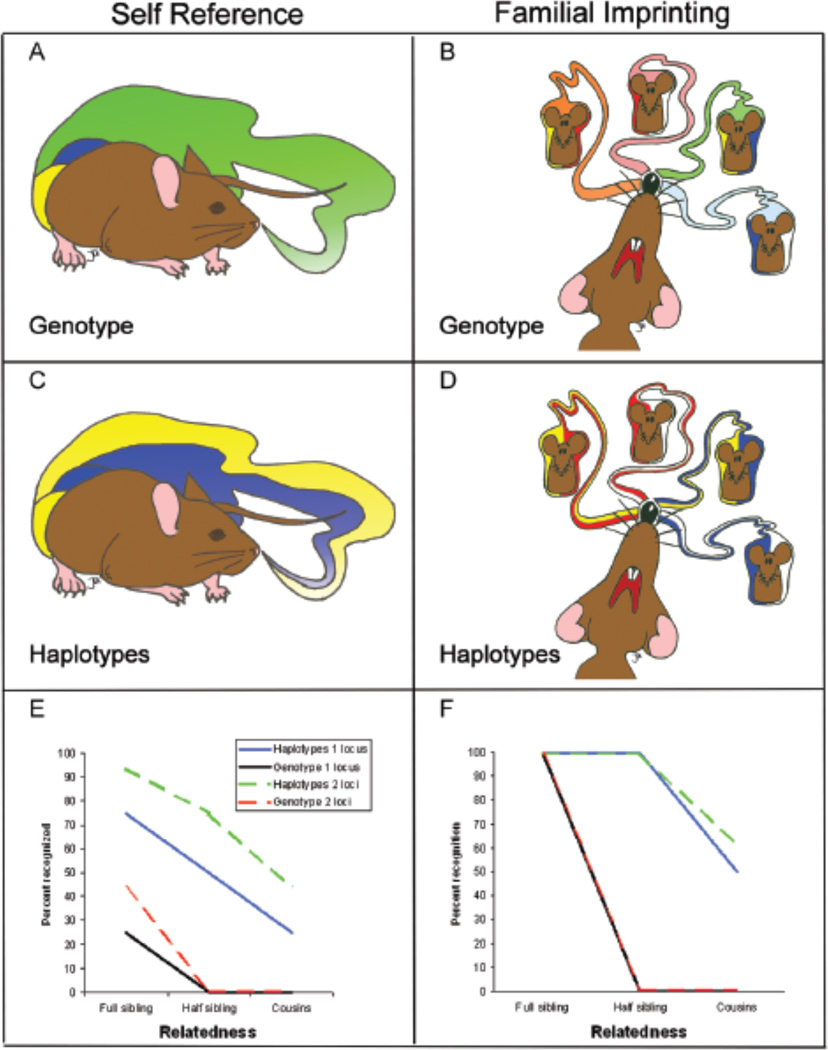
Possible phenotype matching systems using MHC-based odors and their effectiveness for the recognition of kin. Two kin recognition mechanisms that exist in nature are self reference (A, B, E) and familial imprinting (C,D,F). Phenotype matching can be based on haplotypes (e.g. allele specific odors) or on genotypes (e.g. combined haplotype odor). Self-referent phenotype matching can be based on odors associated with genotype (A) or both haplotypes (B). Familial imprinting can be based on odors associated with the genotypes (C) or haplotypes (D) present in the nest (e.g. parents or siblings). The prevalence of these phenotype matching systems in nature is largely untested; current evidence suggests that the primary phenotype matching system in mice is haplotype-based familial imprinting. The effectiveness of each phenotype matching system for recognizing three classes of kin are plotted for one or two unlinked polymorphic loci (E & F). MHC haplotypes contain multiple loci that are inherited in a linked “one locus” system or as multiple unlinked regions “multiple loci” depending on taxa. Models assume that all individuals are heterozygous, that no alleles are shared between unrelated individuals and that all combinations of parental genotypes are found within litters. Haplotype signaling is always superior to genotype signaling and two-locus systems provides an advantage in self-referent systems compared to single-locus systems. (Illustrations by J.L.K; graphic design by Linda Morrison).