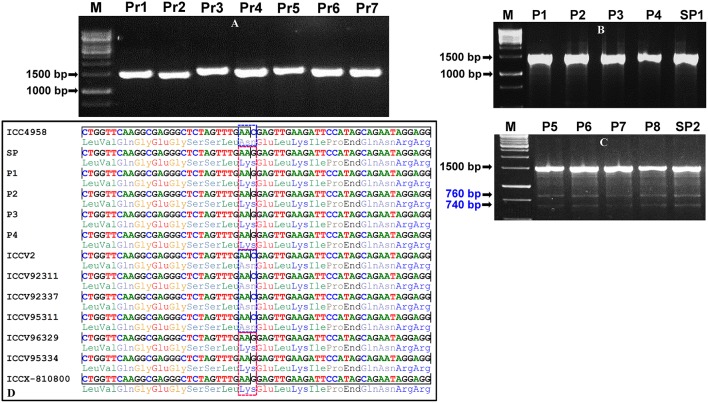
Figure 2.
Optimization and validation of pool-based EcoTILLING approach coupled with agarose gel detection assay for large-scale mining of novel allelic variants from diverse coding and regulatory sequence components of TF genes by genotyping in a 192 desi and kabuli germplasm lines belonging to a seed weight association panel. (A) A representative gel illustrating the optimization followed by PCR amplification of seven primer-pairs (Pr1-Pr7) designed targeting various coding and regulatory regions of seven TF genes in the genomic DNA of a desi chickpea accession (ICC 4958) to produce single reproducible amplicons of each primer for EcoTILLING analysis. (B,C) The representative gels depicting the screening of allelic variants from the eight representative micropools (P1–P8) and two superpools (SP1 and SP2) made from the genomic DNA of 192 desi and kabuli germplasm lines (including ICC 4958 as control) employing an agarose gel-based EcoTILLING assay as defined in the Figure 1. The absence (B) and presence (C) of one non-synonymous SNP allelic variant in the pools and superpools based on cleavage/digestion patterns of 1500 bp fragments amplified from the target CDS region of a mTERF TF gene was apparent in 2.5% agarose gel. The occurrence of 1500 bp homoduplex uncut PCR amplicons as well as mismatch-specific CEL I cleavage of 1500 bp heteroduplex PCR amplified fragments into two varied amplicons of 760 and 740 bp fragment sizes due to the effect of single nucleotide polymorphism (SNP-allele) was observed in the four pools (P5–P8) and one superpool (SP2). M: 1 kb DNA ladder size standard. (D) The sequencing of 1500 bp amplified PCR product of an mTERF TF gene followed by multiple alignment of their high-quality sequences ascertained the presence of one coding SNP (C to G) exhibiting missense non-synonymous amino acid substitution [aspargine (AAC) to lysine (AAG)] in the four pools (P5–P8), one superpool (SP2) and seven individual desi and kabuli accessions as per expectation based on agarose gel-based EcoTILLING assay. The sequenced region carrying the non-synonymous SNP is indicated with a dotted box. The detail information of genes used for validation is mentioned in the Table S2.