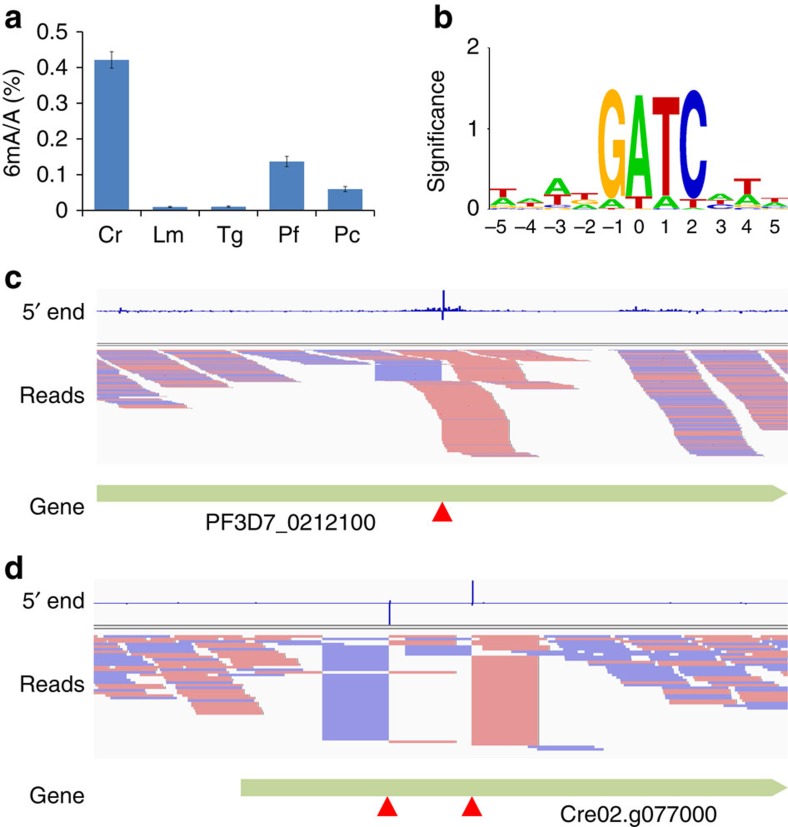
Figure 4. DA-6mA-seq is a highly sensitive method to detect 6mA at specific sites.
(a) Quantification of 6mA abundances in Chlamydomonas reinhardtii (Cr), Leishmania major (Lm), Toxoplasma gondii (Tg), Plasmodium falciparum (Pf) and Penicillium chrysogenum (Pc) by LC–MS/MS. Error bars are calculated as the s.d. of three biological replicates. (b) The accumulated nucleotide composition of all the potential 6mA sites identified by DA-6mA-seq in Plasmodium. Sequence logo is generated by WebLogo (ref. 40). (c) The snapshot of genome browser (IGV (ref. 41)) representing a partially methylated site in Plasmodium. The top track shows counts of 5′ ends in the selected region. The ‘Reads' track shows aligned reads. Blue segments represent reads mapped to minus strand and red segments represent reads mapped to plus strand. The red triangle marks GATC site. (d) The snapshot of genome browser representing two completely methylated sites in Chlamydomonas.