Figure 6. Potential regulation of histone methylation state by histone-modifying enzymes.
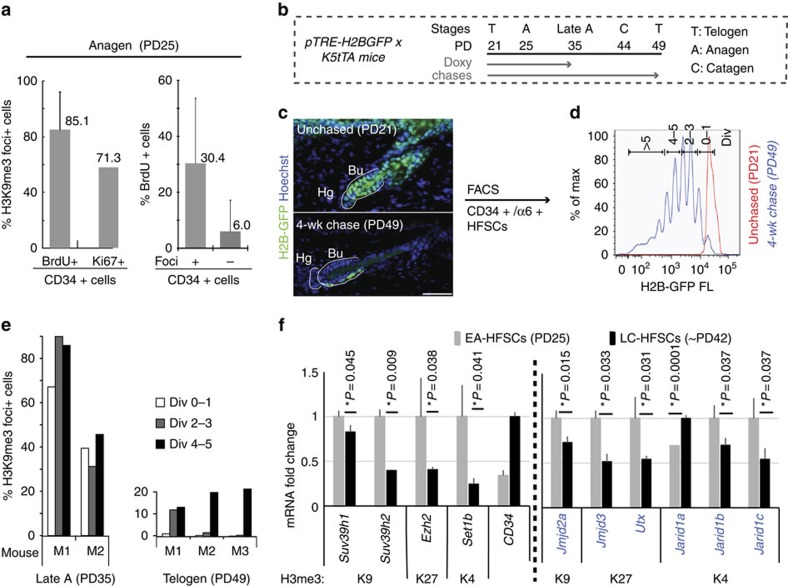
(a) Quantification of foci + and − in CD34+/BrdU+ cells and CD34+/Ki67+ cells (left panel), and % BrdU+ cells in CD34+/foci+ and CD34+/foci− cells. (b) H2B-GFP pulse-chase scheme to examine histone mark level relative to HFSC divisional history. (c) Skin sections and (d) FACS plots of PD21 (unchased) and PD21–49 doxy chased mice. H2B-GFP FACS signal dilutes ½ upon division. Scale bars, 50 μm. (e) % H3K9me3 foci+ in cytospin collection of FACS-isolated HFSCs with different divisional history from late anagen (left) and telogen (right) mice. (f) qRT–PCR of multiple H3 methylases and demethylases in vivo. Statistical significance was analysed using Student's t-test. Refer to Supplementary Table 1 for list of primers used. wk, week.