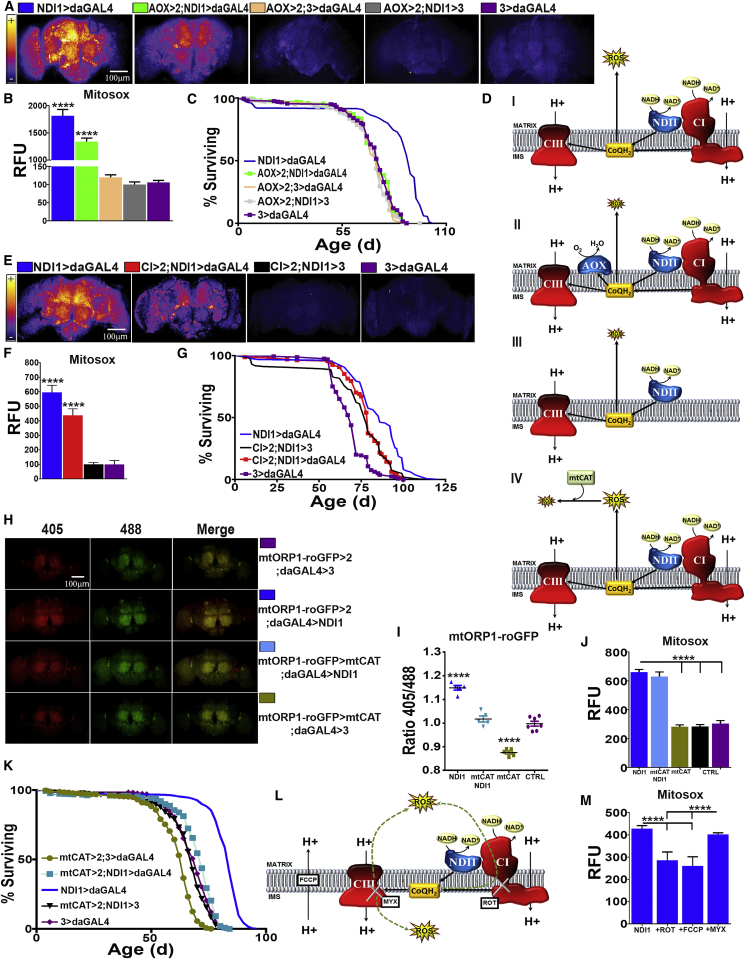
Figure 2.
NDI1 Increases ROS Production via Over-Reduction of CoQ
(A) Representative images of dissected brains from indicated genotypes stained with MitoSOX.
(B) Quantification of (A) (n = 5).
(C) Survival curves for the indicated genotypes (n = 200).
(D) Schematic diagram illustrating effects of expressing two different alternative respiratory enzymes on electron transport: (i) NDI1 generates ROS by over-reducing the CoQ pool; (ii) AOX reverts the effects of NDI1 by re-oxidizing the CoQ pool; (iii) decrease in the levels of CI can prevent reduction of CoQ and subsequent ROS production; (iv) ectopic expression of mtCAT reduces ROS levels without altering mitochondrial respiration or the redox state of CoQ.
(E) Representative images of brains from the indicated genotypes stained with MitoSOX.
(F) Quantification of (E) (n = 5).
(G) Survival curves for the indicated genotypes (n = 160).
(H) In vivo ROS measurements from indicated genotypes in brains dissected from flies expressing a mitochondrially localized redox-sensitive GFP-based reporter.
(I) Quantification of (H) (n = 5–7).
(J) Quantification of brains dissected from flies of the indicated genotypes stained with MitoSOX (n = 5).
(K) Survival curves for the indicated genotypes (n = 200).
(L) Diagram illustrating using metabolic poisons to dissect ROS production: rotenone (ROT), carbonyl cyanide-p-trifluoromethoxyphenylhydrazone (FCCP), or myxothiazol (MYX). Green dashed arrows indicate the possible flow of electrons following CoQ reduction.
(M) Quantification of brains dissected from NDI1 flies fed with metabolic poisons, stained with MitoSOX (n = 4).
Values shown represent means ± SEM of at least 3 biological replicates, unless otherwise stated.
See also Figure S2 and Table S1 for statistical analysis of survival curves.