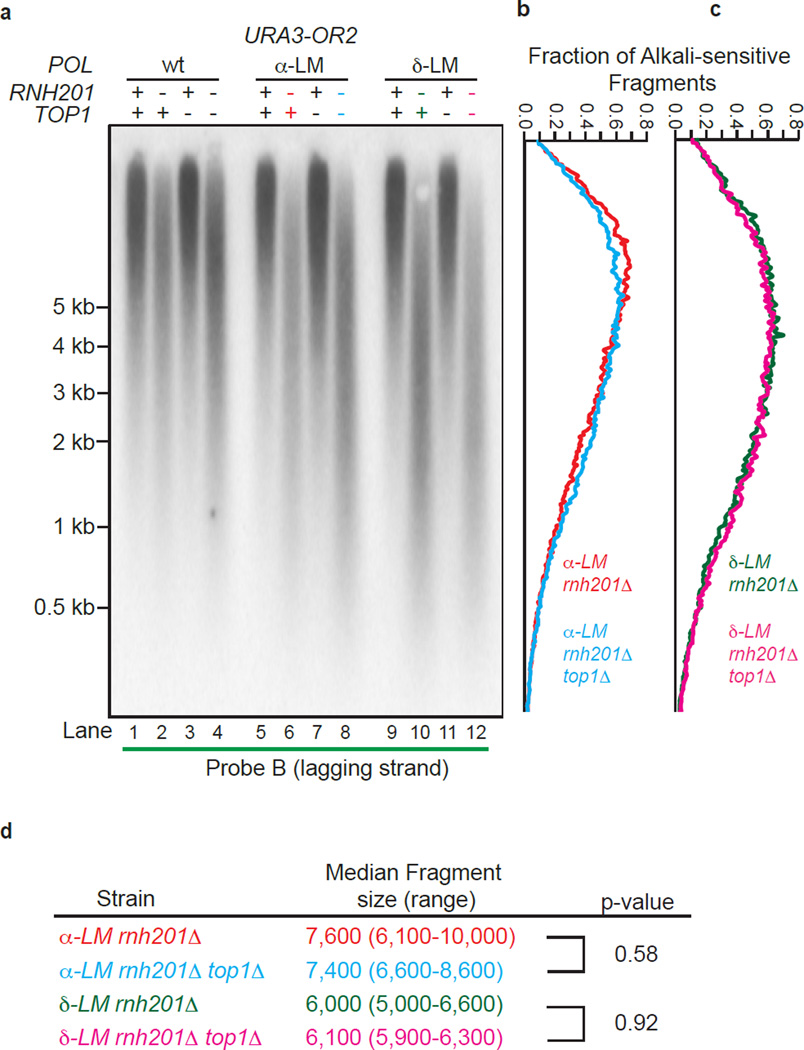
Figure 3.
Lack of Top1-initiated ribonucleotide removal in pol1-L868M rnh201Δ and pol3-L612M rnh201Δ strains. (a) Detection of alkali-sensitive sites in nascent lagging strand DNA was performed using strains that were proficient or deficient in Top1 using the same approach as in Figure 2. All strains contain the URA3 reporter gene in orientation 2 (see Fig. 2a). (b) and (c) The data in panel a were quantified to determine the fraction of total alkali-sensitive fragments at each position along the membrane. The curves are derived using data from three independent experiments. (d) Median DNA fragment sizes (and range) in bases were determined as in13 using quantitation of the alkali-sensitivity data from five (α-LM rnh201Δ and δ-LM rnh201Δ strains) or three (α-LM rnh201Δ top1Δ and δ-LM rnh201Δ top1Δ strains) independent experiments. P values were calculated using a two-tailed Welch’s t-test.