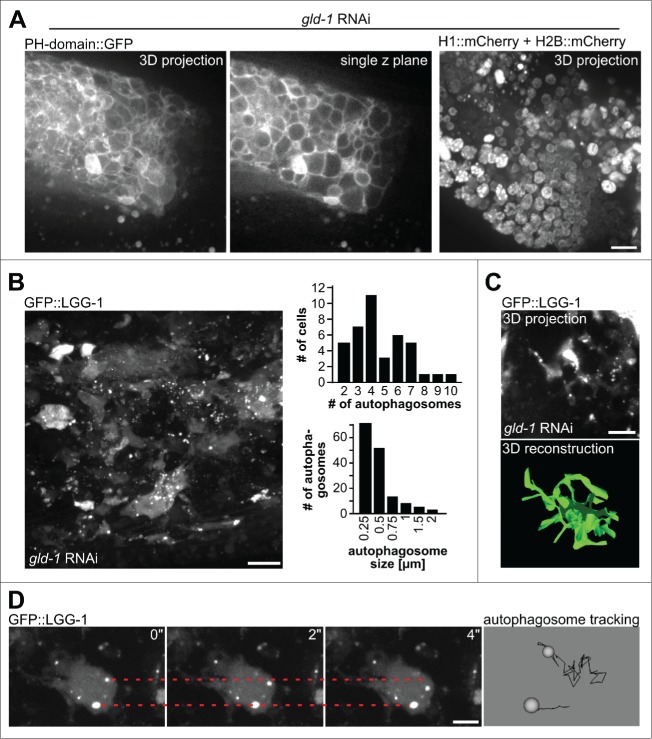
Figure 2.
Characterization of tumor cells and autophagosomes. (A) Tumor cells as visualized by their plasma membranes (left, PH domain of PLCd1 fused to GFP) or their nuclei (histone-mCherry fusion proteins). Scale bar: 10 μm. (B) Left: Autophagosomes in tumor cells as visualized by GFP::LGG-1, a representative maximum projection of a central tumor area is shown, scale bar: 10 μm. Top right: Quantification of numbers of autophagosomes per cell. Bottom right: Quantification of autophagosome size. Quantifications were performed by manual counting/measurements. (C) 3D reconstruction of cell shapes of those cells in the germline tumor expressing GFP::LGG-1. Top: Original microscopy 3D projection data, scale bar: 5 μm; bottom: 3D reconstruction performed in imod (see Methods). (D) Movement of autophagosomes inside tumor cells. Left: Stills from time-lapse recordings, the dashed lines mark the initial position of an autophagosome, scale bar: 5 μm. Right: Magnified data from autophagosome tracking using Endrov (see Methods). Autophagosome paths are shown as black lines. Note that the movement of the bottom autophagosome is due to the sample shifting (see Movie S1). All panels in Fig. 2 show animals at the d 3 of adulthood.