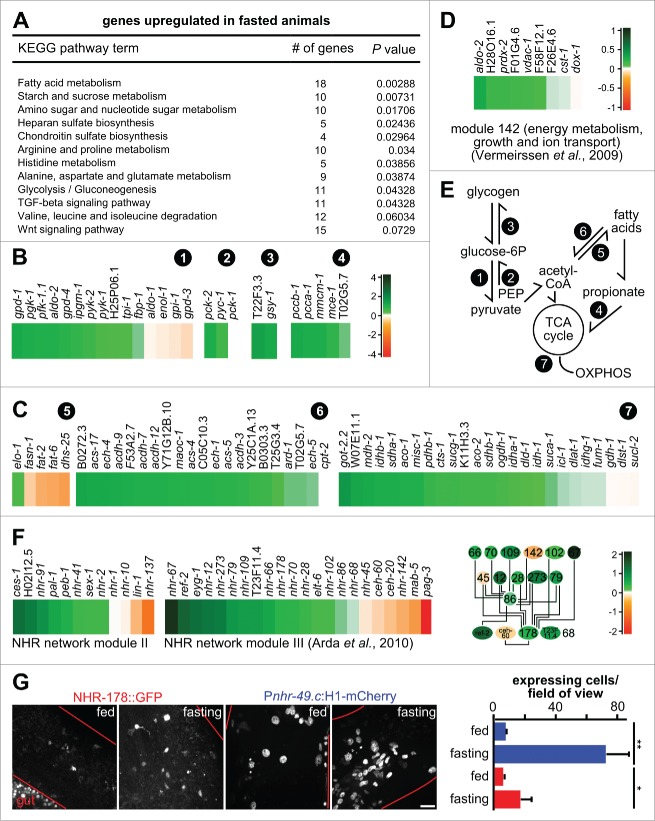
Figure 8.
Fasting induces modular changes in the metabolic gene regulatory network. (A) KEGG pathway enrichment analysis of gld-1 RNAi animals that were fasted. Only upregulated pathways in fasted compared to fed gld-1 RNAi animals with P values lower than 0.01 are shown. (B) Changes in transcript levels in selected carbohydrate and branched-chain amino acid catabolism pathways. log2-fold changes between fasting and feeding are shown. (circled numbers: 1 = glycolysis; 2 = gluconeogenesis; 3 = glycogen metabolism; 4 = propionate metabolism). (C) Changes in transcript levels in fatty acid and mitochondrial metabolic pathways. log2-fold changes between fasting and feeding are shown. log2-scale as in panel (B). (circled numbers: 5 = fatty acid synthesis; 6 = ß-oxidation; 7 = tricarboxylic acid (TCA) cycle, mitochondrial catabolism) (D) Changes in transcript levels in module 142 from ref. 42. log2-fold changes between fasting and feeding are shown. (E) Core metabolic processes in C. elegans and the pathways identified to be regulated after fasting (circled numbers, see above). PEP = phosphoenol-pyruvate, OXPHOS = oxidative phosphorylation. (F) Left: Changes in transcript levels in modules II and III from ref. 45. log2-fold changes between fasting and feeding are shown. Right: Connection between the metabolic regulators in module III. Colors represent log2-fold changes. (G) NHR-178 and NHR-49 expression in germline tumors and regulation by fasting. Left: Representative z-projections of central germline tumor regions of the indicated transgenic animals. Scale bar: 10 μm. Right: Quantification of tumor cells expressing NHR-49 (blue) or NHR-178 (red) at d 3 of adulthood with or without starvation for 1 d; n = 3 animals each; *P ≤ 0.05; **P ≤ 0.01.