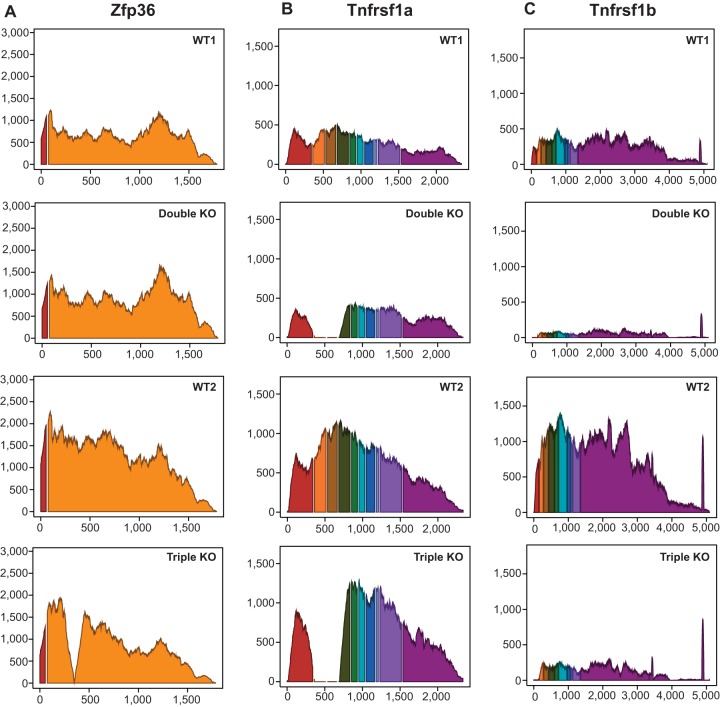
FIG 2.
Average read coverage showing the annotated exons of the three genetically modified transcripts (Zfp36, Tnfrsf1a, and Tnfrsf1b mRNAs) used in the two comparisons. The y axis represents the average sequence read depth at each position for the four samples, and the x axis represents the nucleotide position along the transcript exons, including a fixed 10-nucleotide gap at the site of each intron. Each exon in each transcript is in a different color. (A) Interruption of the reads mapped to the 5′ end of exon 2 of Zfp36, caused by the insertion of the neomycin resistance element, in the triple KO samples. (B) Exons 2 and 3 of Tnfrsf1a were disrupted, as is evident from the absence of reads for those two exons in both the double KO and triple KO samples. (C) Reads were absent from exon 1 in the Tnfrsf1b transcript in both the double and triple KO samples.