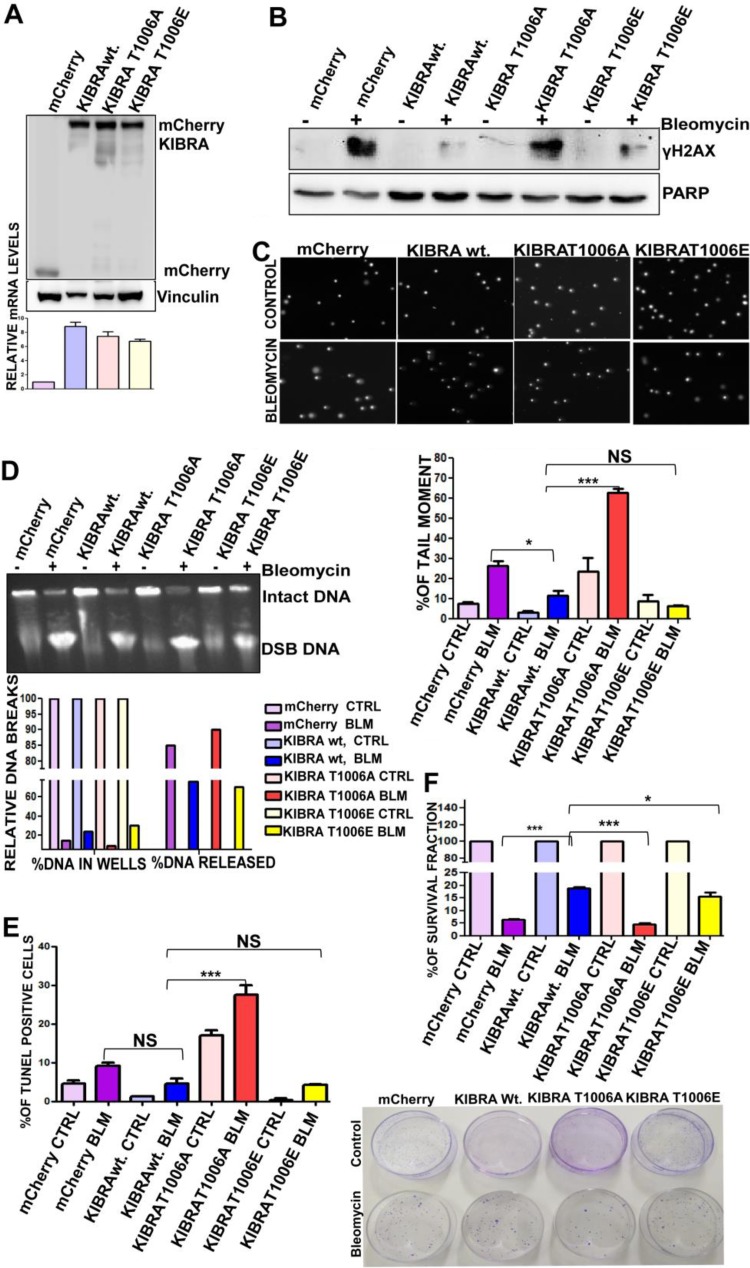
FIG 4.
T1006 phosphorylation on KIBRA is required for optimum DNA double-strand break repair in cancer cells. (A) Western blot and RT-PCR analyses showing mCherry, mCherry-KIBRA-wt., mCherry-KIBRA-T006A, and mCherry-KIBRA-T1006E stable overexpression clones of MCF7 cell line (one-way ANOVA; P < 0.05). (B) MCF7 mCherry, mCherry-KIBRA-wt., and mCherry-KIBRA mutant clones were treated (bleomycin-BLM) or not treated (control-CTRL) with bleomycin for 2 h, and proteins separated using SDS-PAGE followed by blot analysis were probed for γH2Ax (phosphoserine 139); anti-PARP was used as a loading control. (C) Neutral comet assay histogram showing percentages of tail moment of comets, assessed for 100 individual comets using CaspLab software. Percentages of tail moment were compared in mCherry, mCherry-KIBRA-wt., mCherry-KIBRA-T006A, and mCherry-KIBRA-T1006E with or without bleomycin treatment (one-way ANOVA). (D) Percentages of DNA moment from wells with bleomycin treatment or without treatment on MCF7-mCherry, mCherry-KIBRA-wt., and mCherry-KIBRA mutant clones. Undamaged DNA remained in the wells; damaged DNA was released from wells. MCF7-mCherry-KIBRA-wt. and mCherry-KIBRA-T1006E clones were found to have a higher percentage of undamaged DNA. (E) Percentages of survival of MCF7-mCherry, mCherry-KIBRA-wt., and mCherry-KIBRA mutant overexpression clones over 18 days. Cells were seeded at 2,500 cells/plate after the bleomycin treatment for 2 h (one-way ANOVA). (F) Graph showing percentages of terminal deoxynucleotidyltransferase-mediated dUTP-biotin nick end labeling (TUNEL) populations with bleomycin treatment or without treatment in MCF7-mCherry, mCherry-KIBRA-wt., and mCherry-KIBRA mutant clones (one-way ANOVA). *, P < 0.05; ***, P < 0.0005; NS, not significant.